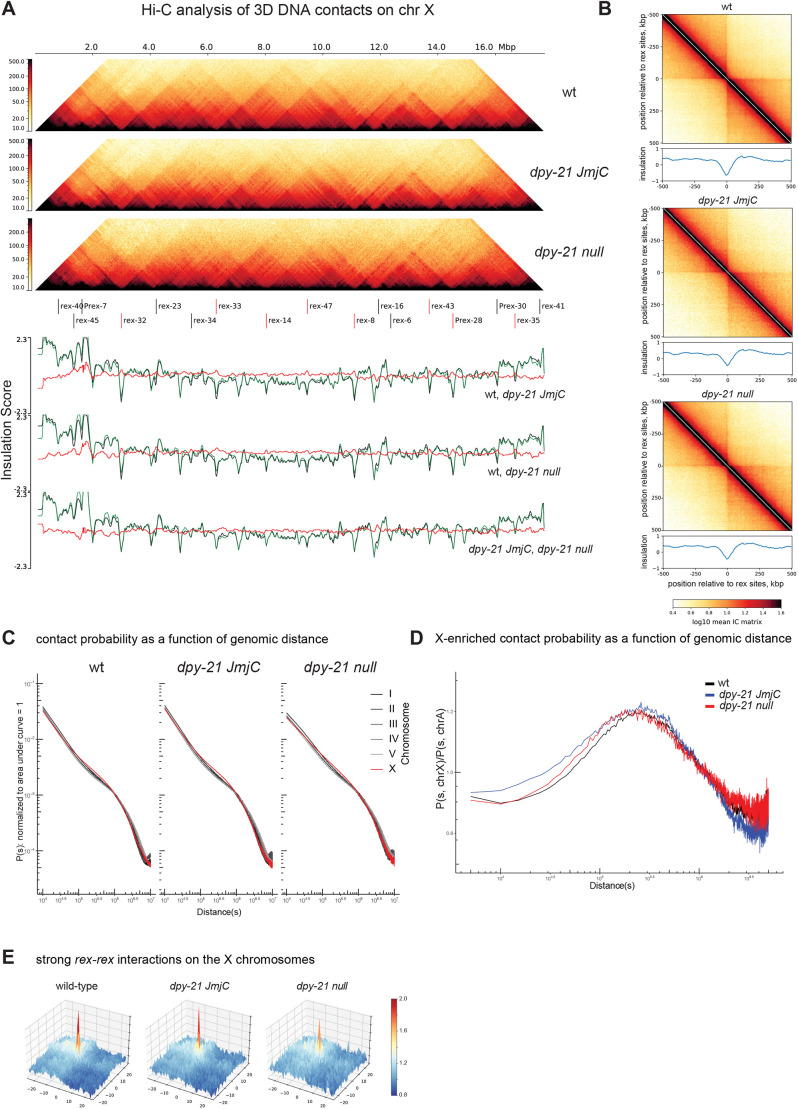
Fig. 5.
Hi-C analysis of 3D DNA contacts in dpy-21(JmjC) and dpy-21 null mutant embryos. (A) Hi-C heatmap (top) and insulation scores (bottom) of the X chromosome (chr X) showing wild type (wt), the dpy-21(JmjC) mutant, and the dpy-21 null mutant. ‘The fall’ colormap, adapted from cooltools, is used to depict the strength of relative contact probability between pairs of genomic bins. The 17 strong rex sites indicated are as annotated by Albritton et al. (2017), eight of which were annotated as DCC-dependent boundary rex sites (indicated by red lines) by Anderson et al. (2019). The insulation scores and their subtractions for three possible pairwise comparisons are shown in the lower panels, The insulation scores for the three pair-wise comparison are as follows: top: wild-type (black), dpy-21(JmjC) (green); middle wild-type (black), dpy-21 null (green); bottom: dpy-21(JmjC) (black), dpy-21 null (green); the red lines indicate per bin subtraction of green minus black. (B) Pile-up analysis showing the average Hi-C map and the insulation scores ±500 kb surrounding the annotated 17 strong rex sites for the indicated genotypes. IC or ‘iteratively corrected' matrix’ is a type of matrix balancing used to correct for different bins having sequencing/representation bias. (C) Distance decay curve showing the relationship between 5-kb binned genomic separation, s, and average contact probability, P(s) computed per chromosome for the indicated genotypes. (D) X chromosome-enriched chromosomal contacts for the indicated genotypes are visualized using an X chromosome minus autosome (X−A)-normalized distance decay curve. For every genomic separation s, the unity-normalized contact probability of the X chromosome, P(s, chrX), is divided by that of autosomes, P(s, chrA). Distances in C and D are shown in bp. (E) Meta-dot plot showing the average strength of interactions between pairs of rex sites on a 10 kb distance-normalized (observed divided by expected) matrix. A total of ±25 bins (±250kb) regions surrounding each rex site are shown. For the 17 strong rex sites, a total of 33 rex–rex pairs located within 3 Mb of each other were used. More blue coloring indicates interaction strengths weaker than expected and more red coloring indicates strength greater than expected.