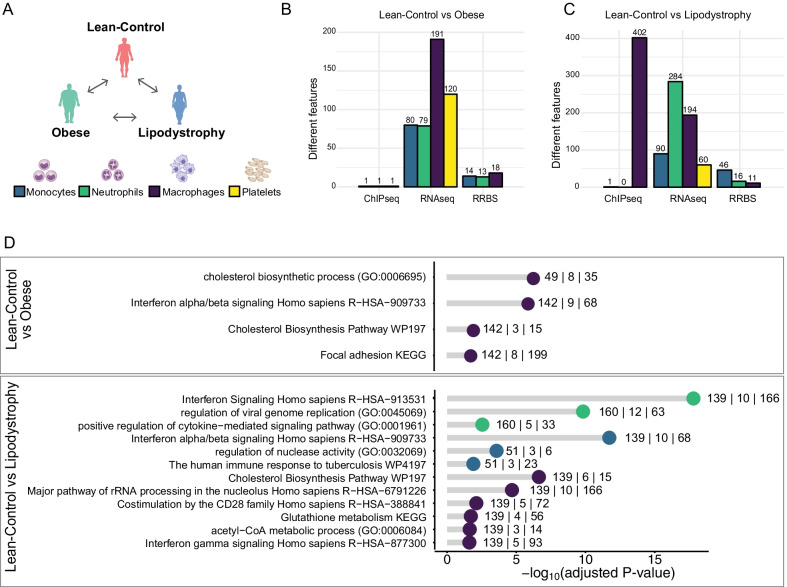
Fig. 2.
Transcriptional and epigenetic signatures in extreme phenotype groups for three innate immune cell types and platelets. a Schematic overview of the comparisons made in the four different cell types (Monocytes: blue; Neutrophils: green; Macrophages: purple; Platelets: yellow). b, c Barplot showing the number of features significantly different: H3K27ac distribution (ChIP-Seq), gene expression (RNA-Seq) and DNA methylation (RRBS). Each bar is colour coded to represent the different cell types as in (a). b represents results when comparing lean-Control and obese individuals. c represents results when comparing lean-Control individuals and lipodystrophy patients. d Functional GO term annotation of upregulated genes when comparing lean-control versus obese group (top) and lean-control individuals versus lipodystrophy group (bottom), colour coded by cell types as above. The numbers near each dot indicate, from left to right: number of submitted genes, number of genes overlapping with the category and number of genes in the category