Fig. 3.
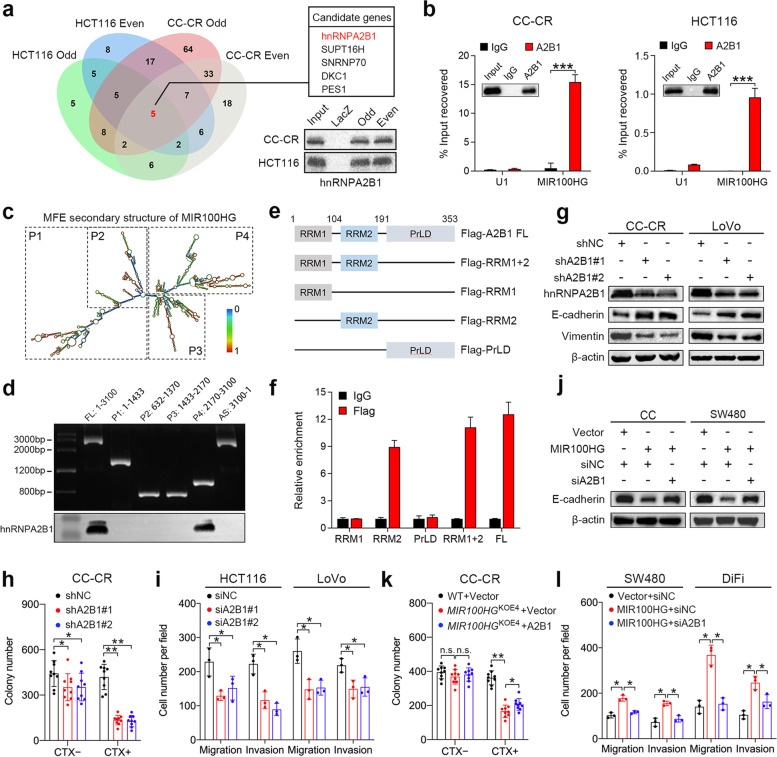
hnRNPA2B1 is a direct and functional binding partner of MIR100HG. a Venn diagram proteins identified by mass spectrometry and retrieved by different groups of MIR100HG probes in ChIRP assays. “Even” and “odd” indicate the two groups of biotin-labeled oilgo probe sets designed for MIR100HG. Immunoblots of hnRNPA2B1 enriched by MIR100HG probes in CC-CR and HCT116 cells (bottom right). b RIP assays detecting MIR100HG retrieved by a hnRNPA2B1 specific antibody or by normal IgG. U1 RNA served as a negative control. Inset depicts immunoblots showing pull-down efficiency. n = 3 independent biological replicates. c Graphic illustration of the predicted secondary structure of MIR100HG. d Top: agarose gel electrophoresis analysis of full-length (FL), truncated or antisense (AS) MIR100HG transcripts; Bottom: immunoblots for hnRNPA2B1 pulled-down by the indicated transcripts of MIR100HG. Representative of three independent experiments. e Diagrams of FL and domain-truncated fragments of hnRNPA2B1. RRM, RNA recognition motif; PrLD, prion-like domain. f Detection of MIR100HG by RIP assay as retrieved by Flag-tagged FL or domain-truncated hnNRNPA2B1 using a Flag antibody. An IgG antibody served as a negative control. n = 3 independent biological replicates. g Immunoblots of indicated EMT markers in CC-CR and LoVo cells after hnRNPA2B1 knockdown. Representative of three independent experiments. h Colony counts of 3D-cultured CC-CR cells after hnRNPA2B1 knockdown in the presence or absence of CTX (3 μg/ml), n = 3 independent experiments performed in triplicate. i Extent of migration and invasion using Transwell migration and invasion assays for indicated cells after hnRNPA2B1 knockdown. n = 3 independent biological replicates. j Immunoblots for E-cadherin in CC and SW480 cells with indicated treatment. Representative of three independent experiments. k Colony counts of 3D-cultured MIR100HGKOE4 CC-CR cells transduced with hnRNPA2B1-expressing lentiviral particles in the presence or absence of CTX (3 μg/ml), n = 3 independent experiments performed in triplicate. l Extent of migration and invasion using Transwell migration and invasion assays for SW480 and DiFi cells with the indicated treatment. n = 3 independent biological replicates. ***P < 0.001, **P < 0.01, *P < 0.05. Data represent mean ± s.d., n.s., not significant