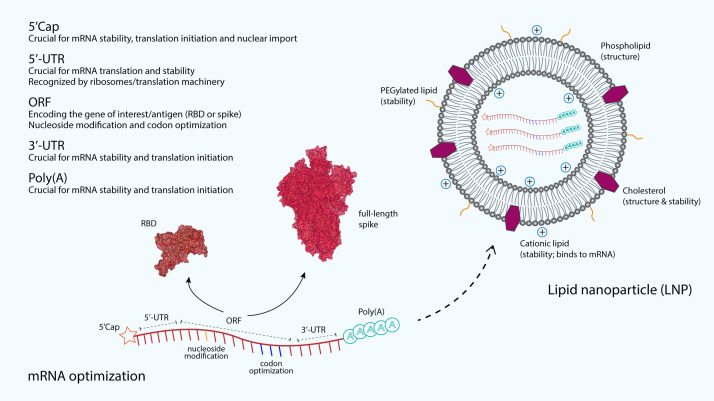
Figure 2. The optimization and delivery of mRNA vaccine.
mRNA vaccines, developed against SARS-CoV-2, are optimized to include a 5′-capping and an untranslated region (UTR) at the 5′-end. The majority of mRNA vaccines developed against SARS-CoV-2 use a m7GpppNmN cap, also known as Cap 1, which is added to the 5′-end using the capping enzyme 2′-O-methyltransferase. These additional structures are added to increase the stability and improve the translation of mRNA. Similarly, an UTR and a poly(A) tail are added at the 3′-end of mRNA, both are crucial for the mRNA stability and translation initiation. The open reading frame (ORF), which encoding the receptor-binding domain (RBD) (PDB: 6M0J) (Lan et al., 2015) or the full-length spike (S) glycoprotein (PDB: 6VSB) (Wrapp et al., 2020), is optimized by using commonly occurring codons to replace rare codons. In addition, the sequence is modified to include GC rich content or modified nucleosides such as 1-methylpseudouridine. mRNA optimization by using modified nucleosides and common codons could potentially increase the immunostimulatory and efficacy of mRNA vaccines. In order to protect mRNA from the extracellular ribonucleases, amine-containing carrier molecules such as lipid nanoparticles (LNPs) are commonly used, which consist of phospholipid bilayer shell and ionizable cationic lipids. In addition, cholesterols and polyethylene glycol (PEG)-containing lipids are also added to increase the overall shell structure and stability.