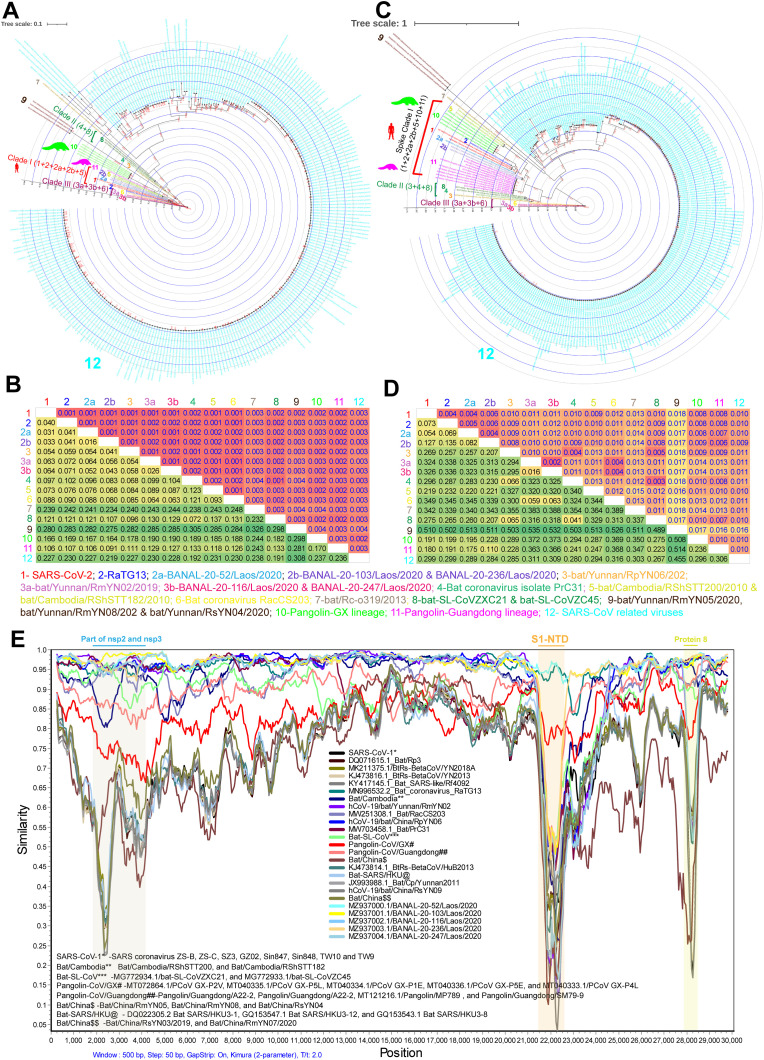
Fig. 1.
The genetic diversity in SARS-CoV-2. (A&B) The Phylogenetic tree (A) and the net between-group mean distance (B), respectively, use the complete genome's nucleotide sequences of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. SARS-CoV-2 related bat viruses n = 18; SARS-CoV-2 related pangolin viruses n = 6; SARS-CoV related viruses n = 258. The different group-specific viruses sequence details are listed in supplementary data 1. (C&D) The Phylogenetic tree (C) and the net between-group mean distance (D) used the complete spike gene nucleotide sequences of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. SARS-CoV-2 related bat viruses n = 18; SARS-CoV-2 related pangolin viruses n = 16; SARS-CoV related viruses n = 256. The different group-specific viruses sequence details are listed in supplementary data 1. (E) The representative Similarity plot is based on the complete genomes nucleotide sequences of the SARS-CoV-2, SARS-CoV-2-related bat/pangolin viruses, and SARS-CoV related viruses. SARS-CoV-2 is used as a reference sequence query. First, we performed the similarity plot analysis with a total of 284 complete genome sequences of SARS-CoV-2 related bat/pangolin viruses and SARS-CoV-related viruses. Next, representative sequences are selected presented here. Details are listed in supplementary data 1.