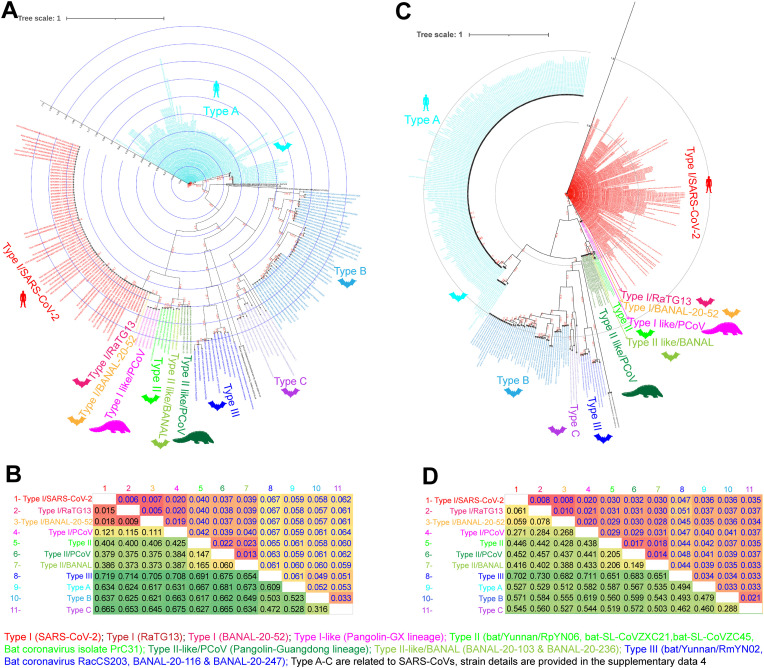
Fig. 4.
The genetic diversity in spike gene of SARS-CoV-2 related viruses. (A&B) The Phylogenetic tree (A) and the net between-group mean distance (B), respectively, use the amino acid sequences of S1-NTDs of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. A total of 236 amino acid sequences of S1-NTDs were used in this analysis. The different Type-specific virus strains details are presented in supplementary data 4. (C&D) The Phylogenetic tree (C) and the net between-group mean distance (D), respectively, use the nucleotide sequences of S1-NTDs of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. A total of 462 nucleotide sequences of S1-NTDs were used in this analysis. The different Type-specific virus strains details are presented in supplementary data 4.