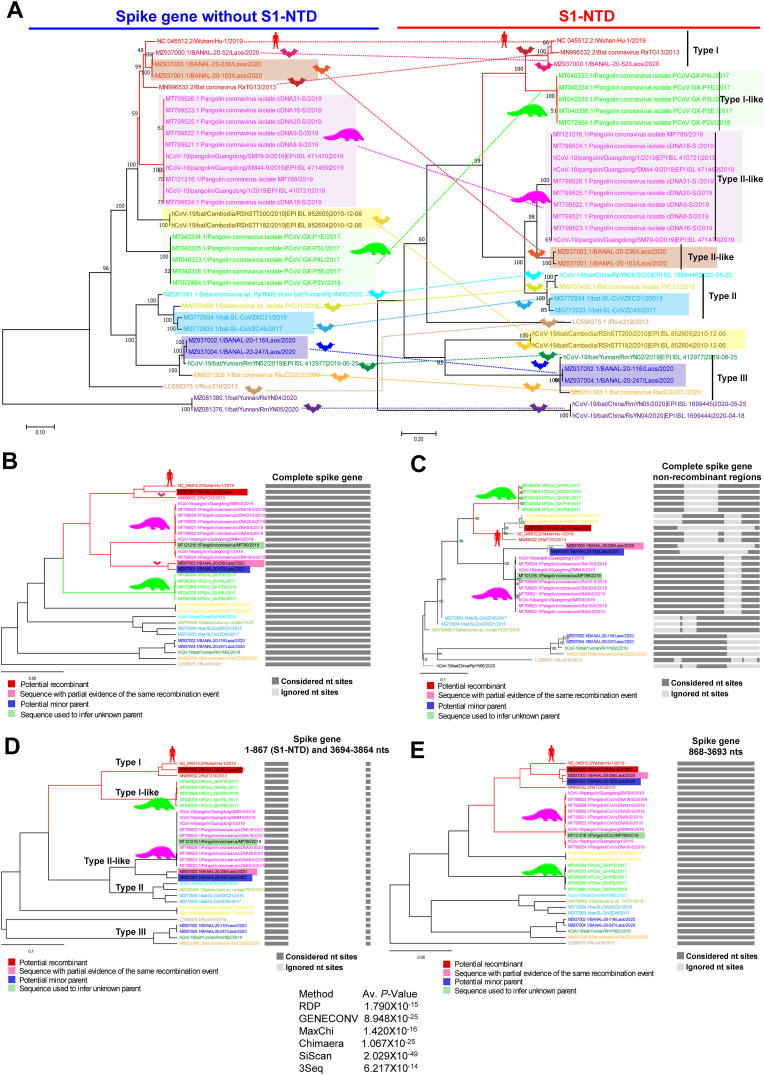
Fig. 5.
Recombination in the spike gene of SARS-CoV-2. (A) Compared the phylogenetic tree using the spike gene without S1-NTD of SARS-CoV-2 related bat/Pangolin viruses with the S1-NTD-based phylogenetic group. The first tree was constructed using SARS-CoV-2 related bat/pangolin viruses spike gene without S1-NTD. The second tree utilized only S1-NTD nucleotide sequences of SARS-CoV-2 related bat/pangolin viruses. (B to E) Phylogenetic tree generated from the RDP using the complete spike sequences of the SARS-CoV-2 related bat/pangolin viruses. The recombination events in the spike gene S1-NTD of SARS-CoV-2 and its related bat/pangolin viruses are supported by RDP, GENECONV, MaxChi, Chimaera, SiScan, and 3Seq. The RDP-based phylogenetic tree using (B) complete spike gene with UPGMA phylogenetic analysis; (C) non-recombinant regions of the entire spike gene with FastNJ phylogenetic analysis; (D) recombinant region of spike gene 1–867 nts (S1-NTD) with UPGMA phylogenetic analysis and 3694–3864 nts; and (E) recombinant region of the spike gene 868–3693 nts with UPGMA phylogenetic analysis.