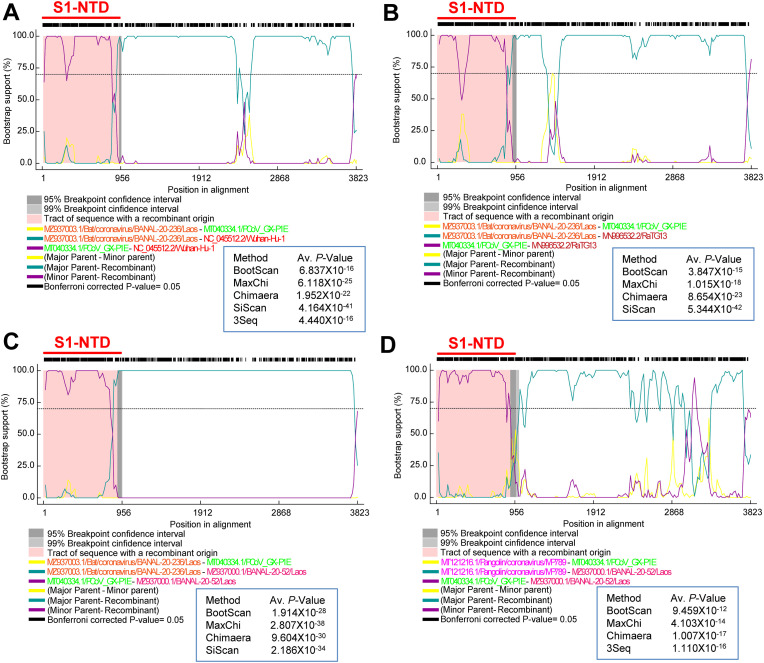
Fig. 6.
Recombination in the Pangolin-Guangdong/BANAL-20-103/BANAL-20-236 spike gene with Pangolin-GX-like Type-I-like-S1-NTD. (A) Representative RDP-based BOOTSCAN depicts the recombination between the BANAL-20-236/Laos and PCoV_GX-P1E in the spike gene. The recombination event is supported by BootScan, MaxChi, Chimaera, SiScan, and 3Seq. It can be inferred that the SARS-CoV-2 spike gene may have been formed by recombining the spike gene of BANAL-20-103/BANAL-20-236 with the S1-NTD of the Pangolin-GX lineage. Because strains BANAL-20-103 and BANAL-20-236 have 98.82% nucleotide sequence identity in the spike gene. Similarly, viruses in the pangolin-GX lineages share 99.5% nucleotide sequence identity in the spike gene. (B) Representative BOOTSCAN depicts the possible origin of the spike gene of RaTG13 by recombining the spike gene of BANAL-20-103/BANAL-20-236 to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. (C) Representative BOOTSCAN depicts the possible origin of the spike gene of BANAL-20-52 by recombining the spike gene of BANAL-20-103/BANAL-20-236 to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. (D) Representative BOOTSCAN depicts the possible origin of the spike gene of BANAL-20-52 by recombining the spike gene of Pangolin-Guangdong to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. Viruses in the pangolin-GX lineages hold 99.5% nucleotide sequence identity in the spike gene. Viruses in the pangolin- Guangdong lineages also share 99.9% nucleotide sequence identity in the spike gene.