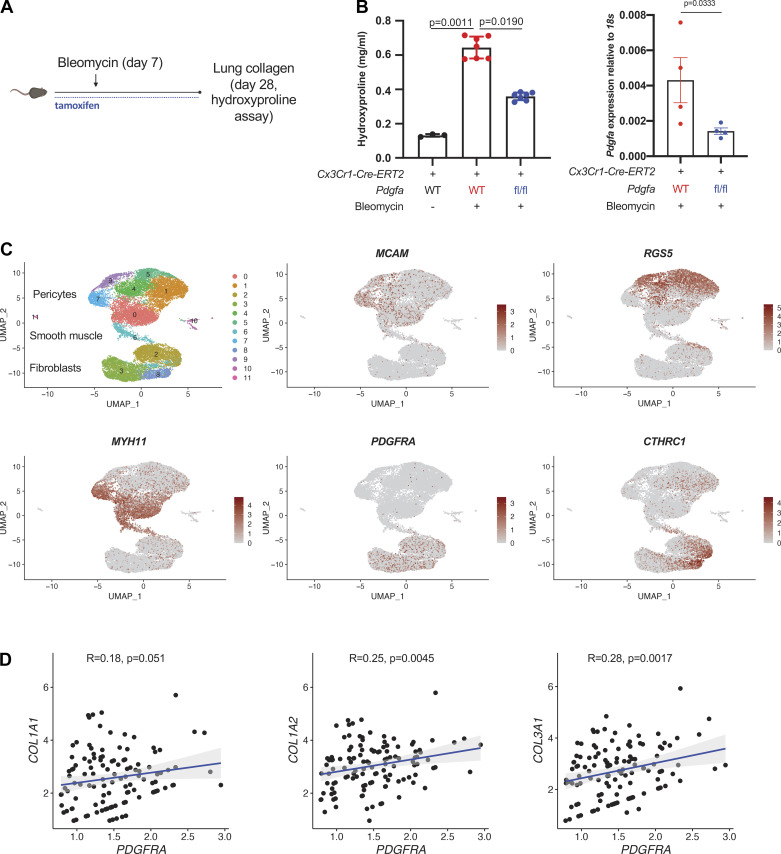
Figure 1.
moMac Pdgfa drives lung fibrosis after bleomycin injury. A: experimental design for inducible, cell type-specific deletion in lung moMacs. B, left: lung hydroxyproline assay in Cx3cr1-CreERT2: Pdgfa fl/fl mice and Cx3cr1-CreERT2 controls (n = 3, 7, and 6 mice/condition as shown). P value is for one-way ANOVA with Dunn’s multiple-comparisons testing; right: qPCR for Pdgfa in macrophages (TdTomato+ cells) sorted from Cx3cr1-CreERT2: Pdgfa fl/fl: R26-loxP-STOP-loxp-TdTomato mice or Cx3cr1-CreERT2: R26-loxP-STOP-loxp-TdTomato controls (n = 4 mice/condition). P value is for one-sided Student’s t test. C: UMAP of scRNA-Seq data for mesenchymal cells from patients with IPF (n = 3 separate individuals) (16). Marker genes shown in feature plots correspond to specific mesenchymal cell types. D: scatter plots show the correlation between PDGFRA and collagen gene expression within the fibroblast compartment; Spearman correlation coefficients and associated P values are shown. IPF, idiopathic pulmonary fibrosis; moMacs, monocyte-derived macrophages; scRNA-Seq, single-cell RNA-Seq. [A was created with BioRender.com.]