Figure 1.
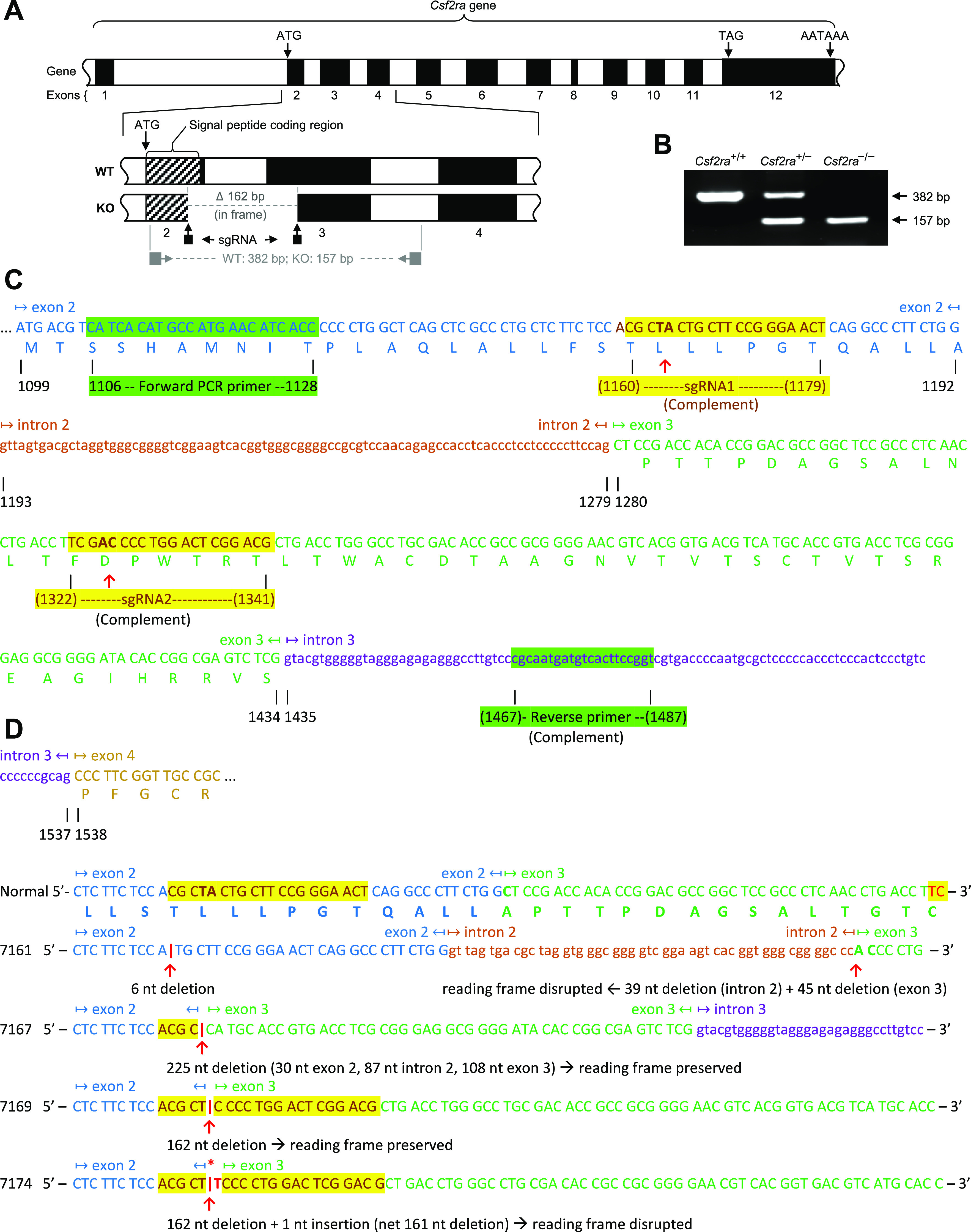
Disruption of Csf2ra gene expression in mice by specific exon-targeted gene editing. A: molecular strategy used to disrupt the Csf2ra gene using CRISPR-Cas9 genome-editing. The normal structure of the Csf2ra gene is shown with the Csf2ra deletion present in mouse line 7169. The location of the single-strand guide ribonucleic acid (sgRNA) and the oligonucleotide primers used for polymerase chain reaction (PCR) amplification-based genotyping of mice are shown. B: PCR-based Csf2ra genotyping analysis showing mice homozygous for wild-type (WT) alleles (Csf2ra+/+), heterozygous for disrupted and WT Csf2ra alleles (Csf2ra+/–), and homozygous for disrupted Csf2ra alleles (Csf2ra–/–). The sizes of normal [382 base pairs (bp)] and disrupted (157 bp) Csf2ra alleles are indicated. C: molecular strategy used to create Csf2ra gene disruption. The sequence of the two sgRNA and oligonucleotide primers used for PCR-based genotyping are shown in alignment to the sequence of mouse chromosome 19 showing a portion of the WT Csf2ra gene [Mus muscularis (C57BL/6J) Chromosome 19, nucleotides 61217191–61228463, mRNA coding strand shown, Genbank accession number NM_009970.2]. D: sequence of the WT Csf2ra gene and the region encompassing exon-2 to exon-3 in each of the four mouse lines (7161, 7167, 7169, and 7174) with disruption of the Csf2ra gene (Csf2ra–/–). The sequences of exons (capital letters) and introns (lower case letters) are indicated, alternating colors are used to improve readability, yellow highlighting identifies the regions affected by gene editing, and the specific deletion in each mouse line is indicated.
