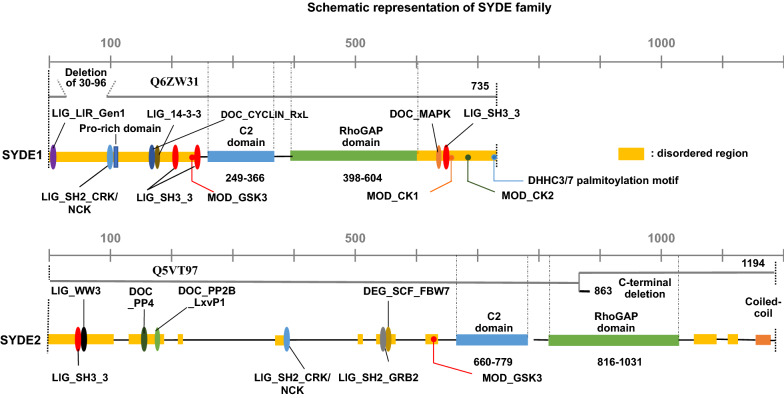
Figure 2.
Domain architecture of SYDE proteins based on their sequence analysis. Both human SYDE proteins are characterized by disorder domain (yellow), C2 (light blue) and RhoGAP domain (green) that are indicated as sequence boundaries of human SYDEs (SYDE1: Q6ZW31; SYDE2: Q5VT97). Conserved linear motifs among vertebrates and mammals for SYDE1 and SYDE2, respectively, are represented as the following according to Dinkel et al. (2016): Canonical LIR motif required for ATG8-mediated autophagy (LIG_LIR_Gen1) (purple oval); Crk and Nck SH2 domain binding motif (LIG_SH2_CRK/NCK) (light blue oval); 14-3-3 interaction motif (LIG_14-3-3) (brown oval); Cyclin/CDK binding motif (DOC_Cyclin_RxL) (dark blue oval); PPR-specific WW domain (LIG_WW3) (black oval); a docking motif mediating interaction with Erk1/2 and p38 subfamilies of MAP kinases (DOC_MAPK) (orange oval); calcineurin docking motif (DOC_PP2B_LxvP1) (light green oval); FxxP-like docking motif recognized by PP4 holoenzyme (DOC_PP4) (dark green); Phsopho-dependent degron recognized by FBW7 Fbox proteins (DEG_SCF_FBW7) (dark yellow oval); Casein kinase (CK1) phosphorylation site (MOD_CK1) (orange dot); CK2 phosphorylation site (MOD_CK2) (dark green dot); DHHC3/7 palmitoylation site (green dot); GSK3 phosphorylation site (MOD_GSK3) (red dot); SH3 binding site (LIG_SH3_3) (red oval); Grb2-like SH2 domains binding motif (LIG_SH2_GRB2) (gray oval). Alternative SYDE isoforms are indicated and the SYDE1 isoform Q6ZW31-2 (668 residues) is missing the N-terminal region (30–96). SYDE2 isoform Q5VT97-2 (863 residues) is truncated at residue 849 with substitution in the region from position 850 to 863 (LSYYGSLLLPLLID).