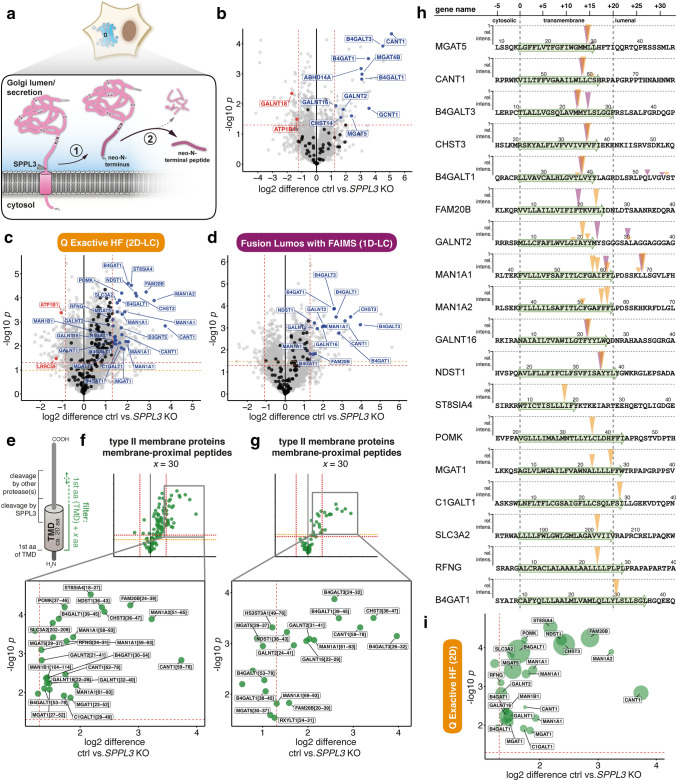
Fig. 2.
N-terminome enrichment approach to identify novel substrates of endogenous SPPL3 and their cleavage sites in CM of isogenic SPPL3-deficient HEK293 cell lines. a Schematic overview of SPPL3-mediated substrate cleavage in the Golgi (blue) and the experimental strategy followed here. Type II membrane protein substrates (pink, with internal trypsin cleavage sites (K/R)) are cleaved by SPPL3 (or other proteases) in the Golgi, are thus liberated from their membrane anchor and secreted (1). Due to proteolysis, liberated cleavage products have free N-terminal amino groups which can be selectively analysed following negative enrichment enabling N-terminome analysis (2). b Label-free quantification of serum-free CM of the three SPPL3-deficient isogenic HEK293 clones and parental HEK293 cells. A total of 3,459 proteins were identified and 131 had their missing values imputed. Proteins derived from type II membrane proteins (based on Uniprot annotations) are displayed in black as well as red or blue, if significantly more abundant in CM of SPPL3 KO or parental cells, respectively. All other proteins identified are given in grey. Red dashed lines represent thresholds applied (log2 difference: median ± 2 SD; p = 0.05 (Welch's t test, two-tail, unequal variance). Gene names are used for labelling. c, d Volcano plots depicting changes in neo-N-terminal peptides enriched via HYTANE from CM of SPPL3-deficient (clone #5 and #6) as well parental cells as measured on the Q Exactive HF system (c, 9,477 unique peptides) and the Fusion Lumos with FAIMS (d 4628 unique peptides). Dashed lines indicate applied thresholds [log2 difference: median ± 2 SD; p = 0.05 (red) and FDR-adjusted p value (orange)]. Volcano plots are filtered for peptides from type II membrane proteins and labelled as in b. e Rationale of peptide filtering. GTfs and other Golgi enzymes adopt a type II membrane topology and their active site containing-ectodomains are anchored to the membrane through a stem region and an N-terminal TMD. Proteolytic cleavage is known to occur in these regions. SPPL3 is expected to cleave type II substrates in the exoplasmic half of the annotated TMD or in a luminal stretch very close to the annotated TMD, other proteases may cleave more distantly from the TMD. Peptides with an N-terminal residue originating in a window comprising the 1st TMD residue + x aa are given in green in subsequent volcano plots. f, g HYTANE datasets shown in c, d but filtered for peptides generated by membrane-proximal cleavage of type II membrane proteins (green). Peptides are labelled with gene name and peptide position (1st aa-last aa). h Schematic mapping of type II membrane protein-derived, TMD-proximal peptides reduced in the CM of SPPL3 KO cells. Protein sequences comprising the TMD and flanking regions were retrieved from Uniprot and aligned at the annotated 1st aa of the TMD (green). Arrowheads indicate cleavage sites (yellow: data from c, purple from d). In the event that multiple membrane-proximal peptides were identified for one protein, height of arrowhead corresponds to the relative intensity of these peptides (mean of three HEK293 replicates). i Data from c, but with point size specifying measured relative peptide intensities (mean of three HEK293 replicates)