Abstract
Primary cilia direct cellular signaling events during brain development and neuronal differentiation. The primary cilium is a dynamic organelle formed in a multistep process termed ciliogenesis that is tightly coordinated with the cell cycle. Genetic alterations, such as ciliary gene mutations, and epigenetic alterations, such as post-translational modifications and RNA processing of cilia related factors, give rise to human neuronal disorders and brain tumors such as glioblastoma and medulloblastoma. This review discusses the important role of genetics/epigenetics, as well as RNA processing and post-translational modifications in primary cilia function during brain development and cancer formation. We summarize mouse and human studies of ciliogenesis and primary cilia activity in the brain, and detail how cilia maintain neuronal progenitor populations and coordinate neuronal differentiation during development, as well as how cilia control different signaling pathways such as WNT, Sonic Hedgehog (SHH) and PDGF that are critical for neurogenesis. Moreover, we describe how post-translational modifications alter cilia formation and activity during development and carcinogenesis, and the impact of missplicing of ciliary genes leading to ciliopathies and cell cycle alterations. Finally, cilia genetic and epigenetic studies bring to light cellular and molecular mechanisms that underlie neurodevelopmental disorders and brain tumors.
Keywords: primary cilia, neurons, epigenetics, RNA splicing, brain tumors
Introduction
The primary cilium is a ubiquitous cellular organelle that resembles an antenna whose main function is to sense extracellular signals through a wide repertoire of receptors present on its surface and linked signaling pathways. Once overlooked as an evolutionary vestige, the cilium has recently emerged as the sensory antenna of many pivotal signaling pathways, such as Sonic Hedgehog (SHH), G-protein coupled receptor (GPCR), platelet-derived growth factor receptor (PDGFR), WNT and TGF-β signaling (Akella et al., 2010; Goetz and Anderson, 2010; Nishimura et al., 2019). Primary cilia play a major role in cellular signaling and are required for SHH, PDGF and WNT signaling cascades that are crucial during neuronal development and patterning. Furthermore, stem cells and neural progenitor cells also possess cilia where they play a critical role in cell cycle, proliferation, and differentiation. Ciliary defects lead to different pathologies collectively termed as ciliopathies. In contrast, many tumor cells lose their primary cilia, resulting in an increase in cell proliferation and malignancy. We will begin presenting some concepts in ciliogenesis and discussing the role of primary cilia in brain development and disease before addressing how post-transcriptional and post-translational modification control cilia status contribute to these processes.
Ciliogenesis
Cilia are microtubule-based structures that emanate from the cell membrane. Whereas motile cilia are found in specialized tissues such as in the respiratory tract and brain ventricles, one primary cilium can be present in every cell. The process to generate a cilium, ciliogenesis, involves several centrosomal proteins and it is tightly coordinated with the cell cycle (Li and Li, 2021). Primary cilia formation can start at G0/G1 phases of the cell cycle, whereas cilia disassembly occurs prior to cell cycle re-entry (Kim and Tsiokas, 2011), and the cell cycle regulation of the primary cilium has been the subject of several detailed reviews (Plotnikova et al., 2009; Kim and Tsiokas, 2011; Goto et al., 2013; Kasahara and Inagaki, 2021; Li and Li, 2021).
Centrosomes are microtubule organizing centers formed by centrioles that are cylindrical structures formed by microtubules that serve as a platform for microtubule polymerization. During mitosis the centrosomes are duplicated and migrate to opposite poles of the cell. After cell division the mother centriole, formed in the previous mitosis, can be dislocated to anchor to the ciliary membrane through the distal appendages. This is an important step to initiate ciliogenesis and, at this point the modified centriole is called a basal body (Kobayashi and Dynlacht, 2011). The basal body is a barrel-like structure where the primary cilia axoneme grows from, with the participation of a specialized transport called intraflagellar transport (IFT) (Ishikawa and Marshall, 2011).
During ciliogenesis ciliary proteins produced in the Golgi can be delivered to the base of the cilium by vesicles as they carry a ciliary target sequence that is recognized by a group of proteins such as Arf4 that regulates the vesicle traffic toward the basal body and the cell membrane. There are other proteins that also participate in this trafficking of vesicles to the cilium such as Rab11, Rab8 and a protein complex called Bbsome that controls IFT assembly and cilia-mediated Hedgehog signaling pathway (Wei et al., 2012; Eguether et al., 2014).
Once vesicles reach the basal body and the cellular membrane, they can be transported along the cilium by IFT proteins that serve as adaptor proteins connecting vesicles to motors that navigate along the axoneme. This process then creates the primary cilium that is a very small and dynamic microdomain in the cell. We will present how primary cilia are involved in different signaling events controlling several processes within the cell such as cell–cell communication, differentiation and cell division (Pugacheva et al., 2007; Gerdes et al., 2009; Goetz and Anderson, 2010; Ishikawa and Marshall, 2011; Kim et al., 2011).
Cilia in Brain Development
Neuronal development is a complex process requiring neural stem cell activity, lineage commitment and neuronal maturation. These events are controlled by signaling pathways that have been shown to be mediated by the primary cilium and ciliary defects are observed in several human genetic syndromes that are characterized by neurological symptoms highlighting the importance of cilia in neuronal development (Table 1).
TABLE 1.
Cilia related genes and epigenetic alterations that have been studied in neurodevelopment and cancer.
| Affected gene | Function | Phenotype | Involved tissue or cells | References | |
| MT PTMs and disease | |||||
| AGTPBP1 (CCP1) | Deglutamylating enzymes | Childhood-onset neurodegeneration with muscular hypotonia, global developmental delays, and cerebellar atrophy | Cerebellum, spinal motor neurons, and peripheral nerves | Magiera et al., 2018 | |
| AGBL5 (CCP5) | Deglutamylating enzymes | Spermatogenesis disruption and male infertility | Spermatids | Giordano et al., 2019 | |
| CCP1 | Deglutamylating enzyme | Upregulated polyglutamylation causes neurodegeneration | Purkinje cells | Rogowski et al., 2010 | |
| Ccp1 and Ccp6 | Deglutamylating enzymes | Excessive polyglutamylation causes neurodegeneration | Cerebral cortex | Shashi et al., 2018 | |
| HDAC6 | Deacetylase | Targeting HDAC6 could be a suitable strategy to ameliorate cognitive decline observed in Alzheimer’s disease | Brain | Govindarajan et al., 2013 | |
| TTLL3 | Tubulin glycylation | Loss of glycylation results in shortening of primary cilia and retinal degeneration | Photoreceptors | Bosch Grau et al., 2017 | |
| TTLL3 | Tubulin glycylation | Loss of glycylation results in loss of primary cilia and cancer formation | Colon | Rocha et al., 2014 | |
| TTLL4 | Tubulin glutamylation | TTLL4 overexpression in breast cancer cells is associated with brain metastasis | Breast and Brain | Arnold et al., 2020 | |
| Primary cilia and neurodevelopmental disorders | |||||
| Arl13b | Ciliogenesis | Axonal tract malformation, Joubert Syndrome Related disorder | Projecting neurons | Guo et al., 2019 | |
| BBS1-19 | Bbsome | BBS, Joubert Syndrome, Meckel-Gruber syndrome, McKusick-Kaufman syndrome, Senior-Loken syndrome | Multisystemic | Novas et al., 2015 | |
| CEP290 | Centrosome | Joubert Syndrome, Meckel-Gruber syndrome, BBS, nephronophthisis, Senior-Loken syndrome | Multisystemic | Coppieters et al., 2010 | |
| CPAP | Centrosome | Seckel Syndrome correlated with microcephaly | Neuronal progenitor | Gabriel et al., 2016 | |
| NEK1 | Cilia assembly | Oral-facial-digital syndrome type II, Mohr Syndrome | Multisystemic | Monroe et al., 2016 | |
| OFD1 | Ciliogenesis | Oral-facial-digital syndrome type I, CNS abnormalities and cystic kidney disease | Multisystemic | Ferrante et al., 2006 | |
| Smo | HH Signaling | Medulloblastoma | Cerebellum | Han et al., 2009 | |
| Alternative splicing of genes encoding cilia proteins in brain tumors | |||||
| CDK20 aka CCRK | Cell cycle, ciliogenesis, development | Various cancers (GBM, medulloblastoma, colorectal, ovarian, hepatocellular, lung, ovarian etc.), developmental defects | Brain tumors, epithelial tumors, embryogenesis, neural patterning, eye, lung, skeletal morphogenesis | Snouffer et al., 2017. | |
| ULK3 | Ciliogenesis, autophagy | Bartter syndrome, autophagy, positive and negative regulator of Shh signaling | Brain, respiratory epithelia | Maloverjan et al., 2010 | |
| STK36 aka Fused | Primary cilia, Hedgehog pathway | Hydrocephalus, primary cilia diskynesia | Ubiquitous, gastric tumors, testis, ovaries | Wilson et al., 2009; Han et al., 2019 | |
| NEK1 | Cilia assembly | Polycystic Kidney Disease, Oral-facial-digital syndrome type II, Mohr Syndrome, ALS | Kidney, CNS, skeletal, multisystemic | Monroe et al., 2016 | |
| Splicing factors essential for ciliogenesis | |||||
| PRPF8 | Spliceosome snRNP formation | Retinitis Pigmentosa | Retina | Tanackovic et al., 2011b; Wheway et al., 2015 | |
| PRPF6 | Spliceosome snRNP formation | Retinitis Pigmentosa | Retina | Tanackovic et al., 2011a; | |
| PRPR31 | Spliceosome snRNP formation | Retinitis Pigmentosa | Retina | Buskin et al., 2018 | |
| SON | Splicing factor | ZTTK multiple congenital anomalies-mental retardation syndrome | Brain | Stemm-Wolf et al., 2021 | |
|
SANS/USH1g |
Ciliary scaffold protein | Usher Syndrome | Inner ear, cochlear hair cell bundles | Yildirim et al., 2021 | |
| SRSF1 | Splicing factor | Cancer, autoimmune disease | Ubiquitous | Haward et al., 2021 | |
| MAGOH | Exon junction complex core factor | Microcephaly neural stem cells | Neural stem cells | Kwon et al., 2021 | |
| RBM8A | Exon junction complex core factor | Thrombocytopenia-Absent radius (TAR) syndrome, microcephaly | Limb development, hematopoiesis, brain development | Kwon et al., 2021 | |
| EIF4A3 | Exon junction complex core factor | Robin sequence, ccenter mandible, and limb anomalies (Richieri-Costa-Pereira syndrome), microcephaly | Craniofacial, limb and brain development | Kwon et al., 2021 | |
| Epigenetic factors involved in ciliogenesis | |||||
| HDAC6 | Epigenetics, histone and tubulin deacetylation | Chondrodysplasia, brachydactyly, hydrocephaly | ubiquitous | Hubbert et al., 2002; Pugacheva et al., 2007; Smith et al., 2018 | |
| HDAC3 | epigenetics, histone deacetylation | Circadian rythms, metabolism, cardiomyopathy | Ubiquitous | Park et al., 2019 | |
| HDAC8 | Epigenetics, histone deacetylation | Cornelia de Lange mental retardation syndrome | Brain | Park et al., 2019 | |
| HDAC2 | Epigenetics, histone deacetylation | Cancer | Ubiquitous | Kobayashi et al., 2017 | |
| SIRT2 | Histone and protein deacetylase | Anoxia, Wallerian degeneration, aging | Ubiquitous | Zhou et al., 2014 | |
| ATAT1 | a-tubulin acetyltransferase | Forebrain development, motor neurons | Brain | Akella et al., 2010; Shida et al., 2010 | |
| WDR5 | Histone modifications, MLL complex | Kcenterrsa syndrome, Kabuki syndrome, center-right asymetry | Ubiquitous | Kulkarni and Khokha, 2018 | |
| EZH2 | H3K27 histone methyltransferase | Weaver syndrome, mantle cell lymphoma, leukemia, breast cancer, melanoma, prostate cancer | Ubiquitous | Zingg et al., 2018 | |
| SMYD2 | Protein methyltransferase | Cancer, cardiovascular | Ubiquitous | Li et al., 2020 | |
| MLL2 | H3K4 methyltransferase | Leukemia, dystonia 28-childhood onset | Ubiquitous | Yang et al., 2019 | |
| G9A/GLP | H3K9 methyltransferase | Embryonic development, cognition-adaptive behavior, obesity | Ubiquitous | Shin et al., 2015 |
Primary cilia participate in neuronal development as they are crucial for early brain patterning, neurogenesis, neuronal maturation, and survival mostly through regulating cell cycle progression and key developmental signaling pathways such as SHH and WNT (Guemez-Gamboa et al., 2014; Youn and Han, 2018). Moreover, severe brain alterations are found in different diseases related to primary cilia disfunction (Table 1). Importantly, alterations and disruption in ciliogenesis dysregulate signaling events that are implicated not only in several neurodevelopmental diseases but also brain tumors, highlighting the importance of cilia in brain development (Table 1).
The SHH signaling pathway guides brain development and SHH signals depend on proper primary cilia IFT machinery to be carried in and out of the cilium. The SHH pathway regulates cell fate and self-renewal, and it is activated in the cilium. In the presence of SHH ligand, the receptor Smoothened is recruited to the cilium promoting the formation of activated forms of GLI (GLIA) that migrate into the nucleus and modulate target genes (Anvarian et al., 2019). It was demonstrated that disruption of SHH signals in IFT mouse mutants lead to neural tube and patterning defects (Huangfu et al., 2003) such as disorganized midline (Gazea et al., 2016).
Besides the SHH signaling pathway that is modulated by the primary cilium, WNT signaling also plays an important role in brain development. The WNT pathway controls cell proliferation, differentiation, migration, apoptosis and tissue morphogenesis (Ng et al., 2019). The involvement of cilia in WNT signaling is not as well studied as SHH but some studies have shown that the protein inversin plays a crucial role in the planar cell polarity (PCP) pathway by recruiting disheveled to the plasma membrane upon WNT receptor Frizzled activation (Simons et al., 2005; Lienkamp et al., 2010). The primary cilium was also shown to act as a regulator of canonical WNT signaling in kif3a knockout embryos that lack cilia and display overactivation of WNT reporter (Corbit et al., 2008).
Cilia, Neural Stem Cells, and Neural Progenitors
Neuronal formation requires proliferation of neuronal stem cells, cellular migration, and differentiation. There are different models of cell division observed in neuronal progenitors such as self-amplifying division to produce two progenitors, self-renewing division to produce one progenitor and one differentiating cell, and self-consuming division to produce two differentiating cells. Importantly, cell proliferation and the exit of the cell cycle are crucial for neurogenesis as it determines the size of the brain and the formation of neuronal and glial cells (Ohnuma and Harris, 2003).
Primary cilia assemble and disassemble during the different phases of the cell cycle functioning as a barrier to cell cycle progression, therefore cilia defects affect proliferation and differentiation of neural progenitors leading to developmental malformations and diseases (Youn and Han, 2018), notably microcephaly and learning disabilities as summarized in Table 1.
Mutations in ciliary proteins are associated with ciliary disruption and cell cycle alterations. Disruption in WDR62 protein, cause of microcephaly in humans (Bilguvar et al., 2010; Bhat et al., 2011; Kousar et al., 2011; Zombor et al., 2019) regulates centriole’s activity and cilia disassembly. WDR62 localizes to the basal body (Zhang et al., 2019) and is involved in IFT88 recruitment to the cilium (Shohayeb et al., 2020) indicating its involvement in ciliogenesis. The importance of WDR62 protein in brain development was studied in murine and human cerebral organoids. WDR62-deficient organoids displayed smaller brain sizes as a result of the depletion of neuronal progenitors (Zhang et al., 2019). Microcephaly was associated with defective NPC population that presented impaired cilia disassembly leading to increased cilia length and defective proliferation and differentiation. Defective cilia length and progenitor proliferation was rescued by Kif2a overexpression highlighting the importance of the ciliary proteins WDR62 and KIF2A in cilia disassembly and NPC maintenance during brain formation (Zhang et al., 2019).
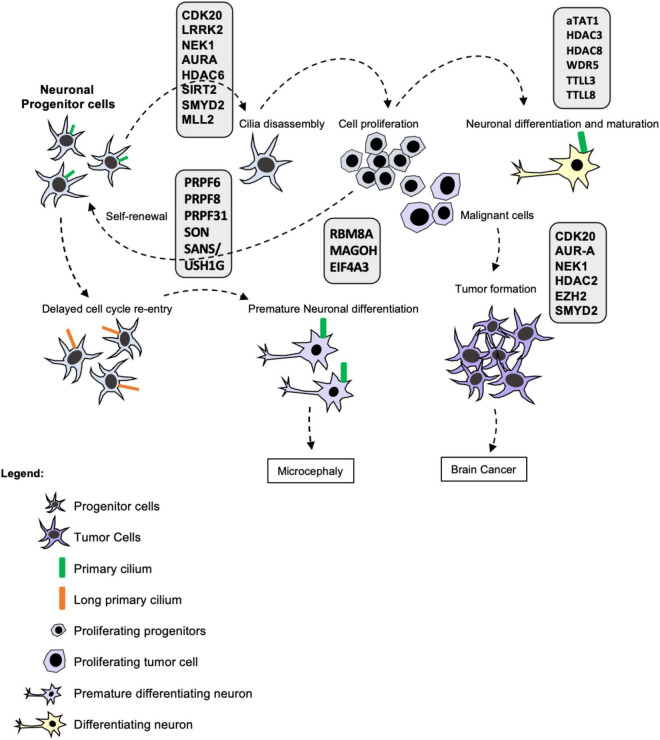
Gabriel et al. (2016) demonstrated that a centrosomal protein called CPAP (centrosomal-P4.1-associated protein) plays a crucial role in cilia disassembly and maintenance of the NPC pool. This work described that CPAP provides a scaffold for the cilia disassembly complex acting as a negative regulator of ciliary length. CPAP is mutated in Seckel syndrome and is correlated with microcephaly. Seckel patient-derived fibroblasts presented long cilia associated with delayed cilia disassembly and cell cycle re-entry (Gabriel et al., 2016). iPSC-derived neuronal progenitor cells (NPCs) from Seckel patients presented signs of early differentiation such as the neuronal marker TUJ1, contained long cilia, reduced proliferative capacity and delayed G1-S transition measured by incorporation of EdU and cyclin A levels (Gabriel et al., 2016). The delay in cell cycle re-entry causes premature differentiation of NPCs reducing the pool of progenitors and ultimately leading to microcephaly (Alcantara and O’Driscoll, 2014; Gabriel et al., 2016). Thus, primary cilia are important to regulate NPC maintenance by controlling the differentiation of neuronal progenitors. Moreover, the primary cilium blocks cell cycle progression inhibiting self-renewal and allowing lineage commitment via neuronal differentiation (Figure 1).
FIGURE 1.
Primary cilia in brain development, maturation, and disease. Ciliogenesis is tightly coordinated with the cell cycle as primary cilia assemble and disassemble during the different phases functioning as a barrier to cell cycle progression. Whereas resting and differentiated cells form a cilium, proliferating cells such as stem cells and progenitor cells disassemble their primary cilia prior to cell division. Stem cell proliferation is important for stem cell self-renewal, maintenance of the progenitor pool and subsequent lineage commitment and differentiation. Many post-transcriptional and post-translational modifications take place in the cilium modulating its formation, stability and activity as depicted in the figure. Therefore, cilia defects and absence affect proliferation and differentiation of neural progenitors leading to developmental diseases and cancer such as Glioblastoma and Medulloblastoma. Cilia disassembly defects cause elongation delaying cell cycle re-entry leading to premature differentiation of NPCs and reducing the pool of progenitors and ultimately leading to microcephaly. Moreover, the ciliary status of neuronal cells from patients with neurodevelopmental and neurodegenerative diseases is yet to be determined. Thus, cilia function is essential for stem cell maintenance, neural development and neural function in health and disease.
Cilia During Lineage Commitment and Differentiation
Primary cilia regulate the patterning and morphogenesis of the forebrain by modulating cellular division and acting as a signaling hub concentrating receptors of morphogens and downstream effectors that guide neuronal differentiation. Mouse studies have shown that mutations in genes involved in IFT that are essential for ciliogenesis and cilia maintenance like Ift72 and Kif3a (Spassky et al., 2008; Gorivodsky et al., 2009) disrupted cell migration and proper formation of neuronal tissue resulting in brain patterning defects. Moreover, these alterations were associated with disrupted Hedgehog signaling showing the importance of primary cilia in modulating SHH-mediated morphogenesis. Similarly, IFT88 alterations cause severe telencephalic architectural disorganization. Ift88 mutations were associated with cranio-facial developmental defects in mice, however, linked to impaired Wnt signaling (Tian et al., 2017).
Ciliopathies and Neurodevelopmental Diseases
Primary cilia play important roles during brain development controlling patterning and morphogenesis of the forebrain. Abnormal cilia formation or function resulting from genetic mutations are called Ciliopathies. Ciliopathies are heterogeneous diseases that can affect almost all organs as primary cilia can be formed in every cell of the human body from stem cells (Yanardag and Pugacheva, 2021), to neurons (Dahl, 1963; Handel et al., 1999; Bishop et al., 2007), epithelial cells in different organs (Jain et al., 2010; Rocha et al., 2014), and blood and bone marrow cells (Singh et al., 2016). Thus, alterations in primary cilia can lead to a range of different phenotypes. Brain alterations observed in ciliopathies include brain malformation, microcephaly, hydrocephalies, corpus callosum agenesis, retinal degeneration, intellectual disability, and autism spectrum disorder as well as brain tumors (Table 1).
Ciliopathies present brain alterations and axonal tract defects are a common feature of human ciliopathies. Joubert Syndrome is a well-studied ciliopathy with known associated genes and characteristic brain malformations such as axonal tract defects. Guo et al. (2019) studied the effect of Arl13b deletion in neurons and observed axonal growth defects in vitro. They demonstrated that convergent signaling pathways such as PI3K, AKT and AC3 originated in the primary cilia are propagated across the neuronal cell body to modulate axonal development (Guo et al., 2019). Interestingly, this work demonstrated that ciliary receptor activation modulates growth cone dynamics and axonal development indicating that primary cilia activity is important for neuronal development and connectivity (Guo et al., 2019).
Primary Cilia in Neurodegenerative Diseases
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases characterized by degeneration of dopamine neurons from the substantia nigra and accumulation of misfolded protein aggregates (Lewy bodies) in the brain (Halliday et al., 2014). LRRK2 (leucine-rich repeat kinase 2) autosomal dominant mutations are a major risk factor for PD (Alessi and Sammler, 2018). LRRK2 is a kinase that phosphorylates a subset of Rab GTPases such as Rab8 and Rab10 that are involved in ciliogenesis (Jeong and Lee, 2020). However, Rab8A and Rab10 have opposing roles in normal ciliogenesis. Rab8A promotes cilia elongation, whereas Rab10 suppresses ciliogenesis as it binds to RILPL1 (Rab interacting lysosomal protein-like 1) protein and hinders cilia formation (Dhekne et al., 2018).
The association of LRRK2 mutation and ciliogenesis disruption was observed in PD patient-derived iPSCs that fail to form a cilium in culture (Dhekne et al., 2018). Moreover, neurons in the striatum of LRRK2 mutant mice displayed reduced number of ciliated cells and expressed lower levels of Gli1 transcripts (Dhekne et al., 2018). It is possible then that these alterations reduce neuronal response to SHH in PD patients carrying LRRK2 mutations. Since the primary cilium plays an important role in the SHH signaling pathway (Goetz and Anderson, 2010; Ho and Stearns, 2021), and SHH signals are required to generate human iPSC-derived midbrain dopaminergic neurons (Cooper et al., 2010), cilia loss due to LRRK2 mutations affects the dopaminergic neuron population.
The importance of LRRK2 in primary cilia formation was further tested in mouse embryonic fibroblast (MEF) cells expressing the pathogenic R1441C LRRK2 mutation. MEFs expressing R1441C LRRK2 at endogenous levels failed to produce a primary cilium, whereas cells treated with LRRK2 inhibitor MLi-2 were able to form cilia (Sobu et al., 2021).
Thus, accumulating evidence has uncovered a functional role for LRRK2 in primary cilia formation in murine and human models. These data highlight a molecular pathway involving cilia and SHH signals for proper neuronal function that is altered in Parkinson’s patients. Strikingly, these data show that primary cilia are important neuronal sensors not only during developmental stages but throughout differentiation and later stages of neuronal maturation and ultimately play an important role in neurodegenerative processes.
Primary Cilia and Brain Tumors
Multiple recent studies have reported defects associated with primary cilia in various cancers (Higgins et al., 2019; Sarkisian and Semple-Rowland, 2019). As primary cilia are important for signaling and cell cycle and are assembled from centrioles that organize spindle poles, it is likely that the absence of the organelle may promote tumorigenesis by aberrant signal transduction and cell cycle regulation (Figure 1). Primary cilia are diminished or lost in multiple cancers, including pancreatic ductal adenocarcinoma (PDAC), renal cell carcinoma, basal cell carcinoma, breast cancer, ovarian cancer, prostate cancer, medulloblastoma, cholangiocarcinoma, melanoma and glioblastoma (reviewed by Higgins et al., 2019; Sarkisian and Semple-Rowland, 2019). Several studies have reported aberrant ciliogenesis in glioblastoma (GBM) lines and primary samples reviewed by Alvarez-Satta and Matheu (2018) and Sarkisian and Semple-Rowland (2019). Interestingly, primary cilia formation was reported to be disrupted at early GBM stages so that fully formed cilia were either completely absent, extremely rare or abnormal after incomplete ciliogenesis (Moser et al., 2009, 2014) indicating that primary cilia are involved in brain tumor pathogenesis.
Are cilia functionally important in glioblastomagenesis? Functional studies revealed that primary cilia loss mediated by cyclin-dependent kinase 20 (CDK20, formerly named as cell-cycle related kinase or CCRK) overexpression promotes GBM proliferation (Yang et al., 2013). CCRK behaves as an oncogene in GBM as depleting CDK20/CCRK inhibited tumor growth (Ng et al., 2007; Yang et al., 2013). Its downregulation also restored ciliogenesis thus indicating that a decrease in cilia promotes tumor growth (Yang et al., 2013). In support of this, Loskutov et al. (2018) also showed that ciliary loss is necessary to maintain a highly proliferative phenotype in GBM. This is not surprising as ciliogenesis is tightly regulated and coordinated with the cell cycle.
Further support for the tumor-suppressing role of cilia in GBM was recently reported by Goranci-Buzhala et al. (2021) who showed that patient-derived glioblastoma stem cells displayed high levels of proteins that are involved in cilia disassembly such as CPAP, Aurora-A, NEK2, NDE1 and OFD1 that resulted in suppressing ciliogenesis. Furthermore, patient-derived glioblastoma stem cells and clinical glioblastoma tissue displayed reduced primary cilia formation associated with increased cell proliferation. Importantly, rescuing ciliogenesis by depleting the disassembly protein complex restored cilia formation and non-malignant phenotype, as glioma stem cells were able to trigger differentiation (Goranci-Buzhala et al., 2021). Strikingly, ciliogenesis-induced differentiation was critical to prevent infiltration of glioma stem cell organoids into the brain of mice showing that signals from the primary cilia determine glioma stem cell fate (Goranci-Buzhala et al., 2021). Collectively, these studies highlight the importance of primary cilia in regulating neuronal differentiation, cell proliferation and, consequently, GBM tumorigenesis as cilia formation is impaired in GBM (Sarkisian and Semple-Rowland, 2019).
Cilia and Medulloblastoma
Medulloblastoma, a tumor arising in the cerebellum, is the most common type of malignant pediatric brain tumor. Medulloblastoma is causally linked to mutations in the SHH pathway that signals through the primary cilium (Rohatgi et al., 2007; Corbit et al., 2008). PTCH1 mutations are the most common MB driver (Northcott et al., 2017). Amplification of the Sonic Hedgehog pathway is observed in the best characterized medulloblastoma subgroup (SHH), with 30% of human tumors having mutations in Patched, Sufu (Suppressor of Fused Homolog), Smoothened, or other genes in this pathway (Northcott et al., 2011, 2017). These mutations lead to constitutively activated SHH signaling, driving tumor development and progression. SHH signaling requires the primary cilium, and both are essential for cerebellar development (Chizhikov et al., 2007; Spassky et al., 2008). SHH is critical for the ontogenesis and patterning of the cerebellum by regulating the expansion and proliferation of neural progenitor cells (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). Notably, cilia are only found in SHH and WNT subgroups but not present in other medulloblastoma subgroups (Han et al., 2009) thus underscoring the functional importance of cilia in tumors that are driven by SHH and WNT signaling. Furthermore, it is worth noting that the SHH and WNT subgroups have better prognosis and outcomes than the other two Medulloblastoma subgroups (Northcott et al., 2011) suggesting that the presence of primary cilia in these tumors results in a less malignant phenotype.
Alternative Splicing of Genes Encoding Cilia Proteins
Alternative splicing affects most human multi-exon genes. As such, alternative splicing has been reported in several genes involved in ciliogenesis. Notably, a recent study looking for splicing changes in GBM subgroups identified enrichment of alternative splicing events in ciliogenesis genes when comparing mesenchymal vs. proneural GBM clusters (Guardia et al., 2020). Expression of some of these genes correlated with prognosis and survival thus underscoring their importance for GBM tumor progression and aggressiveness (Guardia et al., 2020). In our recent study of PRMT5 inhibition in patient-derived GBM stem cells, we identified transcriptome-wide altered splicing (Sachamitr et al., 2021). Notably, we observed enrichment of altered splicing in genes involved in cilia and centrosomes. CDK20 (aka CCRK) splicing was affected and found to shift to the less proliferative isoform (Sachamitr et al., 2021). In addition, we identified splice shifts in the ULK3 and STK36 kinases (listed in Table 1). ULK3 regulates SHH signaling by phosphorylating the Gli transcription factors (Maloverjan et al., 2010). Similarly, STK36 is a fused kinase homolog which also regulates SHH signaling by regulating the Gli transcription factors (Wilson et al., 2009). We also found splicing shifts in NEK1 kinase which is mutated in polycystic kidney disease and is required for ciliogenesis (Shalom et al., 2008; White and Quarmby, 2008). These results suggest that some of the anti-proliferative effects of PRMT5 inhibitors may be mediated by its impact on missplicing of ciliary kinases.
RNA splicing has also been functionally implicated in Medulloblastoma. Dubuc et al. (2012) identified alternative splicing signatures that can accurately discriminate the four different medulloblastoma subgroups with alternative splicing in SHH and group 3 being the most prevalent. The importance of splicing in medulloblastoma development is further underscored by the recent discovery of recurrent hotspot mutations in the non-coding U1snRNA found exclusively in the SHH medulloblastoma subgroup (Suzuki et al., 2019). The U1snRNA mutations inactivate tumor suppressors such as Patched and activate oncogenes like Gli genes thus functionally affecting SHH and ciliary signaling (Suzuki et al., 2019).
Spliceosome Involvement in Ciliary Diseases Like Retinitis Pigmentosa
Heterozygous mutations in the spliceosomal genes PRPF6, PRPF8 and PRPF31 cause retinitis pigmentosa (RP, Tanackovic et al., 2011a; Table 1). Intriguingly, a recent genome-wide RNAi screen revealed these same pre-mRNA splicing factors PRPF6, PRPF8, and PRPF31 to be essential for ciliogenesis (Wheway et al., 2015). Using patient-derived cells from RP patients bearing these mutations, two independent studies revealed generalized spliceosomal defects affecting alternative splicing profiles in these cells (Tanackovic et al., 2011b; Buskin et al., 2018). PRPF31 mutations disrupt splicing of genes encoding splicing factors and ciliary proteins specifically in the retina tissues correlating with the RP phenotypes observed (Buskin et al., 2018). Furthermore, retina photoreceptor cells display unique splicing patterns, which are not found in transcripts from other cell types and are controlled by the Musashi 1 (MSI1) protein (Wheway et al., 2019). This indicates that tissue specific alternative splicing contributes to the tissue-specific ciliopathy phenotypes. Therefore, ciliopathies can emerge either by genetic variants in spliceosomal proteins, or because of variants affecting splicing of specific cilia genes (Wheway et al., 2019).
Three recent papers further cement the link between splicing factors and ciliogenesis. The splicing factor SON was reported to be required for centrosome assembly and ciliogenesis (Stemm-Wolf et al., 2021). The ciliopathy protein SANS/USH1G, mutations of which lead to Usher syndrome, was shown to regulate pre mRNA splicing and interact with splicing factors SON and SF3B1 (Yildirim et al., 2021). SANS is required for the transfer of tri-snRNPs between Cajal bodies and nuclear speckles which are critical for spliceosome assembly. Furthermore, its depletion or pathogenic SANS mutations were shown to affect the splicing of genes important for cell proliferation and Usher syndrome (Yildirim et al., 2021). An additional layer of complexity was recently discovered by Herve LeHir and colleagues who reported that the exon-junction complex (EJC) is required for ciliogenesis in neural stem cells by accumulating to ciliary bodies and recruiting mRNAs important for centrosome organization (Kwon et al., 2021). Mutations or haploinsufficiency of core EJC components Magoh, RBM8A and EIF4A3 result in microcephaly and neurogenesis defects (Silver et al., 2010; Mao et al., 2015, 2016). All this underscores the functional importance of core splicing factors and RNA localization and processing components in ciliopathies and neurogenesis.
Posttranslational Modification Enzymes With a Role in Ciliogenesis Are Linked to Disease
Cilia has a very unique structure as they are formed by nine peripheral pairs of microtubules that are subjected to the activity of posttranslational modifications (PTMs) enzymes (Ishikawa and Marshall, 2011; Janke and Bulinski, 2011). Tubulin undergoes several highly conserved PTMs including acetylation, detyrosination, glutamylation, and glycylation (Janke and Bulinski, 2011). These PTMs accumulate on a subset of microtubules that are long-lived, including those of cilia and centrioles. Interestingly alterations of PTMs levels have been linked to ciliogenesis defects and tumor formation in mouse models and human samples showed a correlation to tumor progression in murine model for colorectal cancer and samples from patients (Rocha et al., 2014).
Alterations in the level of axonemal glutamylation affect SHH signaling as it interferes with the translocation of GLi3 and tethering of Polycystic Kidney Disease 1/2 (PKD1/2) (He et al., 2018) and impairs the entry of Smoothened and Gli3 into the cilia (Hong et al., 2018). Additionally, glutamylation was also associated with cancer. A study screened for genes with correlation for brain metastasis by microarray and showed that TTLL4, a tubulin glutamylase enzyme, overexpression in breast cancer cells is associated with brain metastasis (Arnold et al., 2020).
Tubulin glycylation is specific to cilia (Bre et al., 1994) and glycylation enzymes are required for primary (TTLL3) and motile cilia (TTLL3 and TTLL8) formation, stability and maintenance (Xia et al., 2000; Wloga et al., 2009, 2010; Pathak et al., 2011; Bosch Grau et al., 2013; Rocha et al., 2014). Gadadhar et al. (2017) showed that glycylation accumulates in primary cilia in a length-dependent manner, and alterations of glycylating enzymes TTLL3 and TTLL8 modulates the length of primary cilia in cultured cells (Gadadhar et al., 2021).
Alterations of glycylation are linked to ciliopathies and cancer. Lack of TTLL3 reduces the number of ciliated cells and increases cell proliferation correlating to tumor progression in a mouse model for colorectal cancer (CRC) showing the importance of glycylation in cilia maintenance (Rocha et al., 2014). Analysis of several CRC cell lines also showed decrease of TTLL3 expression (Campbell et al., 2002; Ikegami et al., 2007) and of primary cilia formation (Rocha et al., 2014). Loss of glycylation was also linked to alterations in all ciliated tissues developing the hallmarks of ciliopathies such as infertility (Campbell et al., 2002; Ikegami et al., 2007; Lee et al., 2013; Konno et al., 2016; Gadadhar et al., 2021) and alterations in respiratory tract (Ikegami et al., 2010), retina (Bosch Grau et al., 2017), and brain ventricles (Bosch Grau et al., 2013).
Acetyl-K40, one of the most conserved tubulin PTMs, is the most abundant tubulin PTM in cilia (Akella et al., 2010). Acetyl-K40 marks long-lived microtubules, including those in the axonemes and basal bodies (Piperno and Fuller, 1985). Tubulin acetylation is important for regulating microtubule architecture and maintaining microtubule integrity. The acetyltransferase responsible for this modification is aTAT1. Abnormal levels of this modification are linked to neurological disorders, cancer, heart diseases and other pathological conditions, thereby yielding important therapeutic implications (Li and Yang, 2015).
Histone Deacetylases
HDAC6 is a histone deacetylase that deacetylates the Acetyl-K40 mark in α-tubulin and regulates microtubule-dependent cell motility (Hubbert et al., 2002). Accumulating evidence shows HDAC6 is required for cilium disassembly (Pugacheva et al., 2007). Knockdown of HDAC6 increases the frequency of primary cilia or make them longer, whereas overexpression lead to fewer or shorter cilia (Pugacheva et al., 2007; Yang et al., 2014; Bangs et al., 2015; Ran et al., 2015). HDAC6 also deacetylates cortactin, and this activity also shortens the cilium, by promoting actin polymerization around the base of the primary cilium (Ran et al., 2015). In addition, HDAC6 is involved in autophagy, clearance of misfolded proteins and tau-mediated neurodegeneration (Yu et al., 2016). SIRT2, an NAD-dependent deacetylase, can also deacetylate a-tubulin (North et al., 2003). Furthermore, SIRT2 regulates ciliogenesis and centrosome amplification (Zhou et al., 2014). Overexpression of SIRT2 in renal epithelial cells disrupted cilia formation, causing decreased numbers of cells with cilia and decreased cilia length, while inhibition of SIRT2 activity blocked cilia disassembly during the cell cycle (Zhou et al., 2014). HDAC2 is essential for suppression of cilia formation by positively regulating Aurora A levels in dividing tumor cells (Kobayashi et al., 2017). In contrast, HDAC3 and HDAC8 are required for cilium assembly and elongation (Park et al., 2019; see Table 1).
Methyltransferases
Other PTM enzymes involved in ciliogenesis are the methyltransferases EZH2 and WDR5. Zingg et al. (2018) showed that EZH2 is a melanoma driver by deconstructing the primary cilia through silencing of cilia genes. They showed that gain of EZH2 promotes loss of primary cilia in benign melanocytic lesions (Zingg et al., 2018). In contrast, blockade of EZH2 activity evokes ciliogenesis and cilia-dependent growth inhibition in malignant melanoma (Zingg et al., 2018). Subsequently, loss of cilia enhances pro-tumorigenic WNT/β-catenin signaling, which drives metastatic melanoma in benign cells.
WDR5 regulates left-right patterning and controls ciliogenesis by acting as a scaffold rather than its histone methyltransferase activity (Kulkarni and Khokha, 2018). In addition, WDR5 stabilizes apical actin architecture to promote multiciliated cell formation (Kulkarni et al., 2018). Specifically, WDR5 binds to basal bodies and migrates apically, where F-actin organizes around WDR5. This function was found to be independent to its chromatin modification activity (Kulkarni et al., 2018). Furthermore, the H3K4 methyltransferase MLL2, which is part of the WDR5 complex, suppresses ciliogenesis through regulating actin dynamics and vesicle transport (Yang et al., 2019). Another methyltransferase, SMYD2, was also recently identified as an α-tubulin methyltransferase involved in ciliogenesis (Li et al., 2020). Depletion of SMYD2 led to increased cilia assembly and elongation (Li et al., 2020). In addition, inhibition of the G9A/GLP H3K9 methyltransferase with inhibitor BIX-01294 was found to induce autophagy and promote cilia elongation in human retina pigmented epithelial cells (Shin et al., 2015). Mutations in several methyltransferase enzymes have neuronal phenotypes such as Weaver syndrome for EZH2 (Gibson et al., 2012), Kabuki syndrome for MLL2 (Ng et al., 2010) and Kleefstra syndrome for the EHMT1/GLP1 gene (Kleefstra et al., 2006) underscoring their importance in neuronal development (Table 1).
Future Directions in Cilia Studies Using Induced Pluripotent Stem Cells as Tools for Drug Discovery
Given the role of primary cilia in neuronal differentiation and tumorigenesis, studies investigating the mechanisms underlying ciliogenesis and its impact on neuronal biology will tremendously benefit the development of therapeutic strategies for the treatment of ciliopathies and several neuronal pathologies including both neurodevelopmental and neurodegenerative diseases, as well as brain cancers. induced pluripotent stem cells (iPSCs) offer a unique platform to study human primary cilia formation and activity during neuronal differentiation, maturation and aging as primary cilia play important roles on neuronal tissue patterning, maturation, survival and tumorigenesis by orchestrating and coordinating important developmental signaling pathways such as WNT and SHH (Youn and Han, 2018).
The advantages of using human iPSCs include: (1) to obtain human neuronal cells that are otherwise rare samples and difficult to obtain for research, (2) the opportunity to study developmental processes such as differentiation and maturation in vitro using appropriate culture media, and (3) the possibility to obtain cells from patients allowing the investigation of cellular mechanisms in pathological contexts. Disease-related mutations can also be introduced by CRISPR-Cas9 technology mimicking various patient genetic backgrounds. In this context, large compound libraries can be tested on iPSC-derived cells allowing the screening for compounds that have an impact on ciliogenesis, maintenance and activity. Finally, (4) iPSCs can be used to generate tissue-specific 3D cultures called organoids to model human tissue development and disease. Organoids provide the opportunity to investigate architectural arrangement, cellular signaling events and molecular mechanisms involved in neurodevelopmental and neurodegenerative disorders, including Ciliopathies, using rare patient-derived samples.
Brain organoids can be obtained by cultivating pluripotent stem cells in 3D suspension and are being used to model a great variety of organs including gut, retina, and brain (Hofer and Lutolf, 2021). The generation of brain organoids first described by Lancaster et al. (2013) opened an avenue for the development and modeling of brain-specific tissue allowing the study not only of developmental disorders such as ASDs (Maussion et al., 2019), but also degenerative processes such as PD (Mohamed et al., 2021) and Retinitis Pigmentosa (Lane et al., 2020). Importantly, as described in this review, primary cilia play critical roles during brain development and tumorigenesis. Using brain organoids to study how genetic and epigenetic alterations modulate these processes using patient-derived cells with disease-specific mutations will allow for unprecedented advances in personalized medicine and drug screening for neurological disorders.
Conclusion
The primary cilium plays crucial roles in modulating cellular signaling pathways that are essential for stem cell maintenance and neural development. Interestingly, ciliary defects are observed in developmental and neurodegenerative diseases, and brain cancers such as Glioblastoma and Medulloblastoma. Moreover, neuronal phenotypes such as microcephaly and intellectual disability are a common feature of human ciliopathies. Ciliogenesis is controlled by cellular processes such as the cell cycle, epigenetics factors, post-translational modifications as well as RNA splicing. This regulation has a strong impact on neuronal development as evidenced by the plethora of neuronal phenotypes observed in enzymatic deficiencies and mutations of epigenetics and PTM enzymes as well as RNA processing components. This role seems to be equally important as the more traditional roles of these enzymes on chromatin dynamics, gene expression and posttranscriptional control.
Moreover, a growing body of evidence is unraveling the importance of the primary cilium in neuronal development, activity and maintenance as demonstrated by human iPSCs-based studies strongly implicating cilia in neurodevelopment, neurodegenerative diseases, and brain cancers. This emphasizes the importance of iPSCs studies in understanding human neuronal pathologies as they provide a platform for modeling human neuronal diseases. Thus, understanding the epigenetic and PTM mechanisms of how cilia are formed, maintained and disassembled is fundamental for neuroscience research and drug discovery.
Author Contributions
CR and PP contributed to conception, design, and writing of the review. Both authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This study was supported by Chamandy Foundation (CR). The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (OGI-196), EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510), Janssen, Merck KGaA (aka EMD in Canada and United States), Pfizer, and Takeda.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fncel.2022.809917/full#supplementary-material
References
- Akella J. S., Wloga D., Kim J., Starostina N. G., Lyons-Abbott S., Morrissette N. S., et al. (2010). “MEC-17 is an alpha-tubulin acetyltransferase.”. Nature 467 218–222. 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcantara D., O’Driscoll M. (2014). “Congenital microcephaly.”. Am. J. Med. Genet. C Semin. Med. Genet. 166C 124–139. 10.1002/ajmg.c.31397 [DOI] [PubMed] [Google Scholar]
- Alessi D. R., Sammler E. (2018). “LRRK2 kinase in Parkinson’s disease.”. Science 360 36–37. 10.1126/science.aar5683 [DOI] [PubMed] [Google Scholar]
- Alvarez-Satta M., Matheu A. (2018). “Primary cilium and glioblastoma.”. Ther. Adv. Med. Oncol. 10:1758835918801169. 10.1177/1758835918801169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L. B., Christensen S. T. (2019). “Cellular signalling by primary cilia in development, organ function and disease.”. Nat. Rev. Nephrol. 15 199–219. 10.1038/s41581-019-0116-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J., Schattschneider J., Blechner C., Krisp C., Schluter H., Schweizer M., et al. (2020). “Tubulin Tyrosine Ligase Like 4 (TTLL4) overexpression in breast cancer cells is associated with brain metastasis and alters exosome biogenesis.”. J. Exp. Clin. Cancer Res. 39:205. 10.1186/s13046-020-01712-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs F. K., Schrode N., Hadjantonakis A. K., Anderson K. V. (2015). “Lineage specificity of primary cilia in the mouse embryo.”. Nat. Cell Biol. 17 113–122. 10.1038/ncb3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat V., Girimaji S. C., Mohan G., Arvinda H. R., Singhmar P., Duvvari M. R., et al. (2011). “Mutations in WDR62, encoding a centrosomal and nuclear protein, in Indian primary microcephaly families with cortical malformations.”. Clin. Genet. 80 532–540. 10.1111/j.1399-0004.2011.01686.x [DOI] [PubMed] [Google Scholar]
- Bilguvar K., Ozturk A. K., Louvi A., Kwan K. Y., Choi M., Tatli B., et al. (2010). “Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations.”. Nature 467 207–210. 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop G. A., Berbari N. F., Lewis J., Mykytyn K. (2007). “Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain.”. J. Comp. Neurol. 505 562–571. 10.1002/cne.21510 [DOI] [PubMed] [Google Scholar]
- Bosch Grau M., Gonzalez Curto G., Rocha C., Magiera M. M., Marques Sousa P., Giordano T., et al. (2013). “Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia.”. J. Cell Biol. 202 441–451. 10.1083/jcb.201305041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch Grau M., Masson C., Gadadhar S., Rocha C., Tort O., Marques Sousa P., et al. (2017). “Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration.”. J. Cell Sci. 130 938–949. 10.1242/jcs.199091 [DOI] [PubMed] [Google Scholar]
- Bre M. H., de Nechaud B., Wolff A., Fleury A. (1994). “Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335.”. Cell. Motil. Cytoskeleton 27 337–349. 10.1002/cm.970270406 [DOI] [PubMed] [Google Scholar]
- Buskin A., Zhu L., Chichagova V., Basu B., Mozaffari-Jovin S., Dolan D., et al. (2018). “Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa.”. Nat. Commun. 9:4234. 10.1038/s41467-018-06448-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P. K., Waymire K. G., Heier R. L., Sharer C., Day D. E., Reimann H., et al. (2002). “Mutation of a novel gene results in abnormal development of spermatid flagella, loss of intermale aggression and reduced body fat in mice.”. Genetics 162 307–320. 10.1093/genetics/162.1.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov V. V., Davenport J., Zhang Q., Shih E. K., Cabello O. A., Fuchs J. L., et al. (2007). “Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool.”. J. Neurosci. 27 9780–9789. 10.1523/JNEUROSCI.5586-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O., Hargus G., Deleidi M., Blak A., Osborn T., Marlow E., et al. (2010). “Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid.”. Mol. Cell Neurosci. 45 258–266. 10.1016/j.mcn.2010.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F., Lefever S., Leroy B. P., De Baere E. (2010). CEP290, a gene with many faces: mutation overview and presentation of CEP290base. Hum. Mutat. 31 1097–1108. 10.1002/humu.21337 [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., et al. (2008). “Kif3a constrains beta-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms.”. Nat. Cell Biol. 10 70–76. 10.1038/ncb1670 [DOI] [PubMed] [Google Scholar]
- Dahl H. A. (1963). “Fine structure of cilia in rat cerebral cortex.”. Z Zellforsch Mikrosk Anat. 60 369–386. 10.1007/BF00336612 [DOI] [PubMed] [Google Scholar]
- Dahmane N., Ruiz i Altaba A. (1999). “Sonic hedgehog regulates the growth and patterning of the cerebellum.”. Development 126 3089–3100. 10.1242/dev.126.14.3089 [DOI] [PubMed] [Google Scholar]
- Dhekne H. S., Yanatori I., Gomez R. C., Tonelli F., Diez F., Schule B., et al. (2018). “A pathway for Parkinson’s Disease LRRK2 kinase to block primary cilia and Sonic hedgehog signaling in the brain.”. Elife 7:e40202. 10.7554/eLife.40202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuc A. M., Morrissy A. S., Kloosterhof N. K., Northcott P. A., Yu E. P., Shih D., et al. (2012). “Subgroup-specific alternative splicing in medulloblastoma.”. Acta Neuropathol. 123 485–499. 10.1007/s00401-012-0959-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguether T., San Agustin J. T., Keady B. T., Jonassen J. A., Liang Y., Francis R., et al. (2014). “IFT27 links the BBSome to IFT for maintenance of the ciliary signaling compartment.”. Dev. Cell 31 279–290. 10.1016/j.devcel.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M. I., Zullo A., Barra A., Bimonte S., Messaddeq N., Studer M., et al. (2006). Oral-facial-digital type I protein is required for primary cilia formation and left-right axis specification. Nat. Genet. 38 112–117. 10.1038/ng1684 [DOI] [PubMed] [Google Scholar]
- Gabriel E., Wason A., Ramani A., Gooi L. M., Keller P., Pozniakovsky A., et al. (2016). “CPAP promotes timely cilium disassembly to maintain neural progenitor pool.”. EMBO J. 35 803–819. 10.15252/embj.201593679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar S., Alvarez Viar G., Hansen J. N., Gong A., Kostarev A., Ialy-Radio C., et al. (2021). “Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility.”. Science 371:eabd4914. 10.1126/science.abd4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadadhar S., Dadi H., Bodakuntla S., Schnitzler A., Bieche I., Rusconi F., et al. (2017). “Tubulin glycylation controls primary cilia length.”. J. Cell. Biol. 216 2701–2713. 10.1083/jcb.201612050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazea M., Tasouri E., Tolve M., Bosch V., Kabanova A., Gojak C., et al. (2016). “Primary cilia are critical for Sonic hedgehog-mediated dopaminergic neurogenesis in the embryonic midbrain.”. Dev. Biol. 409 55–71. 10.1016/j.ydbio.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J. M., Davis E. E., Katsanis N. (2009). “The vertebrate primary cilium in development, homeostasis, and disease.”. Cell 137 32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. T., Hood R. L., Zhan S. H., Bulman D. E., Fejes A. P., Moore R., et al. (2012). “Mutations in EZH2 cause Weaver syndrome.”. Am. J. Hum. Genet. 90 110–118. 10.1016/j.ajhg.2011.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano T., Gadadhar S., Bodakuntla S., Straub J., Leboucher S., Martinez G., et al. (2019). Loss of the deglutamylase CCP5 perturbs multiple steps of spermatogenesis and leads to male infertility. J. Cell Sci. 132:jcs226951. 10.1242/jcs.226951 [DOI] [PubMed] [Google Scholar]
- Goetz S. C., Anderson K. V. (2010). “The primary cilium: a signalling centre during vertebrate development.”. Nat. Rev. Genet. 11 331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranci-Buzhala G., Mariappan A., Ricci-Vitiani L., Josipovic N., Pacioni S., Gottardo M., et al. (2021). “Cilium induction triggers differentiation of glioma stem cells.”. Cell Rep. 36:109656. 10.1016/j.celrep.2021.109656 [DOI] [PubMed] [Google Scholar]
- Gorivodsky M., Mukhopadhyay M., Wilsch-Braeuninger M., Phillips M., Teufel A., Kim C., et al. (2009). “Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain.”. Dev. Biol. 325 24–32. 10.1016/j.ydbio.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan N., Rao P., Burkhardt S., Sananbenesi F., Schlüter O. M., Bradke F., et al. (2013). Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer’s disease. EMBO Mol. Med. 5 52–63. 10.1002/emmm.201201923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H., Inoko A., Inagaki M. (2013). “Cell cycle progression by the repression of primary cilia formation in proliferating cells.”. Cell Mol. Life Sci. 70 3893–3905. 10.1007/s00018-013-1302-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia G. D. A., Correa B. R., Araujo P. R., Qiao M., Burns S., Penalva L. O. F., et al. (2020). “Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles.”. NPJ Genom. Med. 5:2. 10.1038/s41525-019-0108-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guemez-Gamboa A., Coufal N. G., Gleeson J. G. (2014). “Primary cilia in the developing and mature brain.”. Neuron 82 511–521. 10.1016/j.neuron.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Otis J. M., Suciu S. K., Catalano C., Xing L., Constable S., et al. (2019). “Primary Cilia Signaling Promotes Axonal Tract Development and Is Disrupted in Joubert Syndrome-Related Disorders Models.”. Dev. Cell 51 759–774e755. 10.1016/j.devcel.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday G. M., Leverenz J. B., Schneider J. S., Adler C. H. (2014). “The neurobiological basis of cognitive impairment in Parkinson’s disease.”. Mov. Disord. 29 634–650. 10.1002/mds.25857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Wang B., Cho Y. S., Zhu J., Wu J., Chen Y., et al. (2019). Phosphorylation of Ci/Gli by fused family kinases promotes hedgehog signaling. Dev. Cell 50:610–626.e4. 10.1016/j.devcel.2019.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. G., Kim H. J., Dlugosz A. A., Ellison D. W., Gilbertson R. J., Alvarez-Buylla A. (2009). “Dual and opposing roles of primary cilia in medulloblastoma development.”. Nat. Med. 15 1062–1065. 10.1038/nm.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel M., Schulz S., Stanarius A., Schreff M., Erdtmann-Vourliotis M., Schmidt H., et al. (1999). “Selective targeting of somatostatin receptor 3 to neuronal cilia.”. Neuroscience 89 909–926. 10.1016/S0306-4522(98)00354-6 [DOI] [PubMed] [Google Scholar]
- Haward F., Maslon M. M., Yeyati P. L., Bellora N., Hansen J. N., Aitken S., et al. (2021). Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. ELife 10:e65104. 10.7554/eLife.65104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Ma X., Xu T., Li Y., Hodge A., Zhang Q., et al. (2018). “Axoneme polyglutamylation regulated by Joubert syndrome protein ARL13B controls ciliary targeting of signaling molecules.”. Nat. Commun. 9:3310. 10.1038/s41467-018-05867-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M., Obaidi I., McMorrow T. (2019). “Primary cilia and their role in cancer.”. Oncol. Lett. 17 3041–3047. 10.3892/ol.2019.9942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E. K., Stearns T. (2021). “Hedgehog signaling and the primary cilium: implications for spatial and temporal constraints on signaling.”. Development 148:dev195552. 10.1242/dev.195552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer M., Lutolf M. P. (2021). “Engineering organoids.”. Nat. Rev. Mater. 2021 1–19. 10.1038/s41578-021-00279-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. R., Wang C. L., Huang Y. S., Chang Y. C., Chang Y. C., Pusapati G. V., et al. (2018). “Spatiotemporal manipulation of ciliary glutamylation reveals its roles in intraciliary trafficking and Hedgehog signaling.”. Nat. Commun. 9:1732. 10.1038/s41467-018-03952-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L., Anderson K. V. (2003). “Hedgehog signalling in the mouse requires intraflagellar transport proteins.”. Nature 426 83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A., et al. (2002). “HDAC6 is a microtubule-associated deacetylase.”. Nature 417 455–458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Ikegami K., Heier R. L., Taruishi M., Takagi H., Mukai M., Shimma S., et al. (2007). “Loss of alpha-tubulin polyglutamylation in ROSA22 mice is associated with abnormal targeting of KIF1A and modulated synaptic function.”. Proc. Natl. Acad. Sci. U S A. 104 3213–3218. 10.1073/pnas.0611547104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Sato S., Nakamura K., Ostrowski L. E., Setou M. (2010). “Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry.”. Proc. Natl. Acad. Sci. U S A. 107 10490–10495. 10.1073/pnas.1002128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W. F. (2011). “Ciliogenesis: building the cell’s antenna.”. Nat. Rev. Mol. Cell Biol. 12 222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Jain R., Pan J., Driscoll J. A., Wisner J. W., Huang T., Gunsten S. P., et al. (2010). “Temporal relationship between primary and motile ciliogenesis in airway epithelial cells.”. Am. J. Respir. Cell Mol. Biol. 43 731–739. 10.1165/rcmb.2009-0328OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C., Bulinski J. C. (2011). “Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions.”. Nat. Rev. Mol. Cell Biol. 12 773–786. 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- Jeong G. R., Lee B. D. (2020). “Pathological Functions of LRRK2 in Parkinson’s Disease.”. Cells 9:2565. 10.3390/cells9122565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K., Inagaki M. (2021). “Primary ciliary signaling: links with the cell cycle.”. Trends Cell Biol. 31 954–964. 10.1016/j.tcb.2021.07.009 [DOI] [PubMed] [Google Scholar]
- Kim J., Dabiri S., Seeley E. S. (2011). “Primary cilium depletion typifies cutaneous melanoma in situ and malignant melanoma.”. PLoS One 6:e27410. 10.1371/journal.pone.0027410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Tsiokas L. (2011). “Cilia and cell cycle re-entry: more than a coincidence.”. Cell Cycle 10 2683–2690. 10.4161/cc.10.16.17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T., Brunner H. G., Amiel J., Oudakker A. R., Nillesen W. M., Magee A., et al. (2006). “Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome.”. Am. J. Hum. Genet. 79 370–377. 10.1086/505693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Dynlacht B. D. (2011). “Regulating the transition from centriole to basal body.”. J. Cell Biol. 193 435–444. 10.1083/jcb.201101005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nakazono K., Tokuda M., Mashima Y., Dynlacht B. D., Itoh H. (2017). “HDAC2 promotes loss of primary cilia in pancreatic ductal adenocarcinoma.”. EMBO Rep. 18 334–343. 10.15252/embr.201541922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A., Ikegami K., Konishi Y., Yang H. J., Abe M., Yamazaki M., et al. (2016). “Ttll9-/- mice sperm flagella show shortening of doublet 7, reduction of doublet 5 polyglutamylation and a stall in beating.”. J. Cell Sci. 129 2757–2766. [DOI] [PubMed] [Google Scholar]
- Kousar R., Hassan M. J., Khan B., Basit S., Mahmood S., Mir A., et al. (2011). “Mutations in WDR62 gene in Pakistani families with autosomal recessive primary microcephaly.”. BMC Neurol. 11:119. 10.1186/1471-2377-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S. S., Khokha M. K. (2018). “WDR5 regulates left-right patterning via chromatin-dependent and -independent functions.”. Development 145:dev159889. 10.1242/dev.159889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S. S., Griffin J. N., Date P. P., Liem K. F., Khokha M. K. (2018). “WDR5 Stabilizes Actin Architecture to Promote Multiciliated Cell Formation.”. Dev. Cell 46 595–610e593. 10.1016/j.devcel.2018.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O. S., Mishra R., Safieddine A., Coleno E., Alasseur Q., Faucourt M., et al. (2021). “Exon junction complex dependent mRNA localization is linked to centrosome organization during ciliogenesis.”. Nat. Commun. 12:1351. 10.1038/s41467-021-21590-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A., Renner M., Martin C. A., Wenzel D., Bicknell L. S., Hurles M. E., et al. (2013). “Cerebral organoids model human brain development and microcephaly.”. Nature 501 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A., Jovanovic K., Shortall C., Ottaviani D., Panes A. B., Schwarz N., et al. (2020). “Modeling and Rescue of RP2 Retinitis Pigmentosa Using iPSC-Derived Retinal Organoids.”. Stem Cell Rep. 15 67–79. 10.1016/j.stemcr.2020.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. S., He Y., Dougherty E. J., Jimenez-Movilla M., Avella M., Grullon S., et al. (2013). “Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility.”. J. Biol. Chem. 288 15167–15180. 10.1074/jbc.M113.453936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. X., Li X. (2021). “Epigenetically Mediated Ciliogenesis and Cell Cycle Regulation, and Their Translational Potential.”. Cells 10:1662. 10.3390/cells10071662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. X., Zhou J. X., Wang X., Zhang H., Harris P. C., Calvet J. P., et al. (2020). “Cross-talk between CDK4/6 and SMYD2 regulates gene transcription, tubulin methylation, and ciliogenesis.”. Sci. Adv. 6:eabb3154. 10.1126/sciadv.abb3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yang X. J. (2015). “Tubulin acetylation: responsible enzymes, biological functions and human diseases.”. Cell Mol. Life Sci. 72 4237–4255. 10.1007/s00018-015-2000-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienkamp S., Ganner A., Boehlke C., Schmidt T., Arnold S. J., Schafer T., et al. (2010). “Inversin relays Frizzled-8 signals to promote proximal pronephros development.”. Proc. Natl. Acad. Sci. U S A. 107 20388–20393. 10.1073/pnas.1013070107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loskutov Y. V., Griffin C. L., Marinak K. M., Bobko A., Margaryan N. V., Geldenhuys W. J., et al. (2018). “LPA signaling is regulated through the primary cilium: a novel target in glioblastoma.”. Oncogene 37 1457–1471. 10.1038/s41388-017-0049-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiera M. M., Bodakuntla S., Žiak J., Lacomme S., Marques Sousa P., Leboucher S., et al. (2018). Excessive tubulin polyglutamylation causes neurodegeneration and perturbs neuronal transport. EMBO J. 37:e100440. 10.15252/embj.2018100440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloverjan A., Piirsoo M., Michelson P., Kogerman P., Osterlund T. (2010). “Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway.”. Exp. Cell Res. 316 627–637. 10.1016/j.yexcr.2009.10.018 [DOI] [PubMed] [Google Scholar]
- Mao H., McMahon J. J., Tsai Y. H., Wang Z., Silver D. L. (2016). “Haploinsufficiency for Core Exon Junction Complex Components Disrupts Embryonic Neurogenesis and Causes p53-Mediated Microcephaly.”. PLoS Genet. 12:e1006282. 10.1371/journal.pgen.1006282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Pilaz L. J., McMahon J. J., Golzio C., Wu D., Shi L., et al. (2015). “Rbm8a haploinsufficiency disrupts embryonic cortical development resulting in microcephaly.”. J. Neurosci. 35 7003–7018. 10.1523/JNEUROSCI.0018-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe G. R., Kappen I. F., Stokman M. F., Terhal P. A., van den Boogaard M.-J. H., Savelberg S. M., et al. (2016). Compound heterozygous NEK1 variants in two siblings with oral-facial-digital syndrome type II (Mohr syndrome). Eur. J. Hum. Genet. 24 1752–1760. 10.1038/ejhg.2016.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maussion G., Rocha C., Bernard G., Beitel L. K., Durcan T. M. (2019). “Patient-Derived Stem Cells, Another in vitro Model, or the Missing Link Toward Novel Therapies for Autism Spectrum Disorders?”. Front. Pediatr. 7:225. 10.3389/fped.2019.00225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed N. V., Sirois J., Ramamurthy J., Mathur M., Lepine P., Deneault E., et al. (2021). “Midbrain organoids with an SNCA gene triplication model key features of synucleinopathy.”. Brain Commun. 3:fcab223. 10.1093/braincomms/fcab223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J. J., Fritzler M. J., Rattner J. B. (2009). “Primary ciliogenesis defects are associated with human astrocytoma/glioblastoma cells.”. BMC Cancer 9:448. 10.1186/1471-2407-9-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J. J., Fritzler M. J., Rattner J. B. (2014). “Ultrastructural characterization of primary cilia in pathologically characterized human glioblastoma multiforme (GBM) tumors.”. BMC Clin. Pathol. 14:40. 10.1186/1472-6890-14-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L. F., Kaur P., Bunnag N., Suresh J., Sung I. C. H., Tan Q. H., et al. (2019). “WNT Signaling in Disease.”. Cells 8:826. 10.3390/cells8080826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. B., Bigham A. W., Buckingham K. J., Hannibal M. C., McMillin M. J., Gildersleeve H. I., et al. (2010). “Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome.”. Nat. Genet. 42 790–793. 10.1038/ng.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S. S., Cheung Y. T., An X. M., Chen Y. C., Li M., Li G. H., et al. (2007). “Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma.”. J. Natl. Cancer Inst. 99 936–948. 10.1093/jnci/djm011 [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Kasahara K., Shiromizu T., Watanabe M., Inagaki M. (2019). “Primary Cilia as Signaling Hubs in Health and Disease.”. Adv. Sci. 6:1801138. 10.1002/advs.201801138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North B. J., Marshall B. L., Borra M. T., Denu J. M., Verdin E. (2003). “The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase.”. Mol. Cell 11 437–444. 10.1016/S1097-2765(03)00038-8 [DOI] [PubMed] [Google Scholar]
- Northcott P. A., Buchhalter I., Morrissy A. S., Hovestadt V., Weischenfeldt J., Ehrenberger T., et al. (2017). “The whole-genome landscape of medulloblastoma subtypes.”. Nature 547 311–317. 10.1038/nature22973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott P. A., Korshunov A., Witt H., Hielscher T., Eberhart C. G., Mack S., et al. (2011). “Medulloblastoma comprises four distinct molecular variants.”. J. Clin. Oncol. 29 1408–1414. 10.1200/JCO.2009.27.4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novas R., Cardenas-Rodriguez M., Irigoín F., Badano J. L. (2015). Bardet-Biedl syndrome: is it only cilia dysfunction? FEBS Lett. 589 3479–3491. 10.1016/j.febslet.2015.07.031 [DOI] [PubMed] [Google Scholar]
- Ohnuma S., Harris W. A. (2003). “Neurogenesis and the cell cycle.”. Neuron 40 199–208. 10.1016/S0896-6273(03)00632-9 [DOI] [PubMed] [Google Scholar]
- Park S. A., Yoo H., Seol J. H., Rhee K. (2019). “HDAC3 and HDAC8 are required for cilia assembly and elongation.”. Biol. Open 8:bio043828. 10.1242/bio.043828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak N., Austin C. A., Drummond I. A. (2011). “Tubulin tyrosine ligase-like genes ttll3 and ttll6 maintain zebrafish cilia structure and motility.”. J. Biol. Chem. 286 11685–11695. 10.1074/jbc.M110.209817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Fuller M. T. (1985). “Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms.”. J. Cell Biol. 101 2085–2094. 10.1083/jcb.101.6.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova O. V., Pugacheva E. N., Golemis E. A. (2009). “Primary cilia and the cell cycle.”. Methods Cell Biol. 94 137–160. 10.1016/S0091-679X(08)94007-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A. (2007). “HEF1-dependent Aurora A activation induces disassembly of the primary cilium.”. Cell 129 1351–1363. 10.1016/j.cell.2007.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran J., Yang Y., Li D., Liu M., Zhou J. (2015). “Deacetylation of alpha-tubulin and cortactin is required for HDAC6 to trigger ciliary disassembly.”. Sci. Rep. 5:12917. 10.1038/srep12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha C., Papon L., Cacheux W., Marques Sousa P., Lascano V., Tort O., et al. (2014). “Tubulin glycylases are required for primary cilia, control of cell proliferation and tumor development in colon.”. EMBO J. 33 2247–2260. 10.15252/embj.201488466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski K., van Dijk J., Magiera M. M., Bosc C., Deloulme J.-C., Bosson A., et al. (2010). A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 143 564–578. 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Scott M. P. (2007). “Patched1 regulates hedgehog signaling at the primary cilium.”. Science 317 372–376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Sachamitr P., Ho J. C., Ciamponi F. E., Ba-Alawi W., Coutinho F. J., Guilhamon P., et al. (2021). “PRMT5 inhibition disrupts splicing and stemness in glioblastoma.”. Nat. Commun. 12:979. 10.1038/s41467-021-21204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisian M. R., Semple-Rowland S. L. (2019). “Emerging Roles of Primary Cilia in Glioma.”. Front. Cell Neurosci. 13:55. 10.3389/fncel.2019.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom O., Shalva N., Altschuler Y., Motro B. (2008). “The mammalian Nek1 kinase is involved in primary cilium formation.”. FEBS Lett. 582 1465–1470. 10.1016/j.febslet.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Shashi V., Magiera M. M., Klein D., Zaki M., Schoch K., Rudnik-Schöneborn S., et al. (2018). Loss of tubulin deglutamylase CCP1 causes infantile-onset neurodegeneration. EMBO J. 37:e100540. 10.15252/embj.2018100540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H., Kim P. S., Kim E. S., Park S. J., Jo Y. K., Hwang J. J., et al. (2015). “BIX-01294-induced autophagy regulates elongation of primary cilia.”. Biochem. Biophys. Res. Commun. 460 428–433. 10.1016/j.bbrc.2015.03.050 [DOI] [PubMed] [Google Scholar]
- Shida T., Cueva J. G., Xu Z., Goodman M. B., Nachury M. V. (2010). The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U.S.A. 107 21517–21522. 10.1073/pnas.1013728107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohayeb B., Ho U., Yeap Y. Y., Parton R. G., Millard S. S., Xu Z., et al. (2020). “The association of microcephaly protein WDR62 with CPAP/IFT88 is required for cilia formation and neocortical development.”. Hum. Mol. Genet. 29 248–263. 10.1093/hmg/ddz281 [DOI] [PubMed] [Google Scholar]
- Silver D. L., Watkins-Chow D. E., Schreck K. C., Pierfelice T. J., Larson D. M., Burnetti A. J., et al. (2010). “The exon junction complex component Magoh controls brain size by regulating neural stem cell division.”. Nat. Neurosci. 13 551–558. 10.1038/nn.2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Gloy J., Ganner A., Bullerkotte A., Bashkurov M., Kronig C., et al. (2005). “Inversin, the gene product mutated in nephronophthisis type II, functions as a molecular switch between Wnt signaling pathways.”. Nat. Genet. 37 537–543. 10.1038/ng1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Chaudhry P., Merchant A. A. (2016). “Primary cilia are present on human blood and bone marrow cells and mediate Hedgehog signaling.”. Exp. Hematol. 44 1181–1187e1182. 10.1016/j.exphem.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouffer A., Brown D., Lee H., Walsh J., Lupu F., Norman R., et al. (2017). Cell Cycle-Related Kinase (CCRK) regulates ciliogenesis and Hedgehog signaling in mice. PLoS Genet. 13:e1006912. 10.1371/journal.pgen.1006912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Q., Macklin B., Chan X. Y., Jones H., Trempel M., Yoder M. C., et al. (2018). Differential HDAC6 activity modulates ciliogenesis and subsequent mechanosensing of endothelial cells derived from pluripotent stem cells. Cell Rep. 24 895–908.e6. 10.1016/j.celrep.2018.06.083 [DOI] [PubMed] [Google Scholar]
- Sobu Y., Wawro P. S., Dhekne H. S., Yeshaw W. M., Pfeffer S. R. (2021). “Pathogenic LRRK2 regulates ciliation probability upstream of tau tubulin kinase 2 via Rab10 and RILPL1 proteins.”. Proc. Natl. Acad. Sci. U S A. 118:e2005894118. 10.1073/pnas.2005894118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N., Han Y. G., Aguilar A., Strehl L., Besse L., Laclef C., et al. (2008). “Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool.”. Dev. Biol. 317 246–259. 10.1016/j.ydbio.2008.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A. J., O’Toole E. T., Sheridan R. M., Morgan J. T., Pearson C. G. (2021). “The SON RNA splicing factor is required for intracellular trafficking structures that promote centriole assembly and ciliogenesis.”. Mol. Biol. Cell 32:ar4. 10.1091/mbc.E21-06-0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kumar S. A., Shuai S., Diaz-Navarro A., Gutierrez-Fernandez A., De Antonellis P., et al. (2019). “Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma.”. Nature 574 707–711. 10.1038/s41586-019-1650-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G., Ransijn A., Ayuso C., Harper S., Berson E. L., Rivolta C. (2011a). “A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa.”. Am. J. Hum. Genet. 88 643–649. 10.1016/j.ajhg.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G., Ransijn A., Thibault P., Abou Elela S., Klinck R., Berson E. L., et al. (2011b). “PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa.”. Hum. Mol. Genet. 20 2116–2130. 10.1093/hmg/ddr094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Feng J., Li J., Ho T. V., Yuan Y., Liu Y., et al. (2017). “Intraflagellar transport 88 (IFT88) is crucial for craniofacial development in mice and is a candidate gene for human cleft lip and palate.”. Hum. Mol. Genet. 26 860–872. 10.1093/hmg/ddx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace V. A. (1999). “Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum.”. Curr. Biol. 9 445–448. 10.1016/S0960-9822(99)80195-X [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R. J., Scott M. P. (1999). “Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog.”. Neuron 22 103–114. 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Wei Q., Zhang Y., Li Y., Zhang Q., Ling K., Hu J. (2012). “The BBSome controls IFT assembly and turnaround in cilia.”. Nat. Cell Biol. 14 950–957. 10.1038/ncb2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway G., Lord J., Baralle D. (2019). “Splicing in the pathogenesis, diagnosis and treatment of ciliopathies.”. Biochim. Biophys. Acta Gene Regul. Mech. 1862:194433. 10.1016/j.bbagrm.2019.194433 [DOI] [PubMed] [Google Scholar]
- Wheway G., Schmidts M., Mans D. A., Szymanska K., Nguyen T. T., Racher H., et al. (2015). “An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes.”. Nat. Cell Biol. 17 1074–1087. 10.1038/ncb3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. C., Quarmby L. M. (2008). “The NIMA-family kinase, Nek1 affects the stability of centrosomes and ciliogenesis.”. BMC Cell Biol. 9:29. 10.1186/1471-2121-9-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. W., Nguyen C. T., Chen M. H., Yang J. H., Gacayan R., Huang J., et al. (2009). “Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis.”. Nature 459 98–102. 10.1038/nature07883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., Gaertig J. (2010). “Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell.”. Eukaryot Cell 9 184–193. 10.1128/EC.00176-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Webster D. M., Rogowski K., Bre M. H., Levilliers N., Jerka-Dziadosz M., et al. (2009). “TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia.”. Dev. Cell 16 867–876. 10.1016/j.devcel.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Xia L., Hai B., Gao Y., Burnette D., Thazhath R., Duan J., et al. (2000). “Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila.”. J. Cell Biol. 149 1097–1106. 10.1083/jcb.149.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanardag S., Pugacheva E. N. (2021). “Primary Cilium Is Involved in Stem Cell Differentiation and Renewal through the Regulation of Multiple Signaling Pathways.”. Cells 10:1428. 10.3390/cells10061428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Hao H., Wu X., Guo S., Liu Y., Ran J., et al. (2019). “Mixed-lineage leukemia protein 2 suppresses ciliary assembly by the modulation of actin dynamics and vesicle transport.”. Cell Discov. 5:33. 10.1038/s41421-019-0100-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ran J., Liu M., Li D., Li Y., Shi X., et al. (2014). “CYLD mediates ciliogenesis in multiple organs by deubiquitinating Cep70 and inactivating HDAC6.”. Cell Res. 24 1342–1353. 10.1038/cr.2014.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Roine N., Makela T. P. (2013). “CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner.”. EMBO Rep. 14 741–747. 10.1038/embor.2013.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim A., Mozaffari-Jovin S., Wallisch A. K., Schafer J., Ludwig S. E. J., Urlaub H., et al. (2021). “SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes.”. Nucleic Acids Res. 49 5845–5866. 10.1093/nar/gkab386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn Y. H., Han Y. G. (2018). “Primary Cilia in Brain Development and Diseases.”. Am. J. Pathol. 188 11–22. 10.1016/j.ajpath.2017.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Ran J., Zhou J. (2016). “Ciliopathies: Does HDAC6 Represent a New Therapeutic Target?”. Trends Pharmacol. Sci. 37 114–119. 10.1016/j.tips.2015.11.002 [DOI] [PubMed] [Google Scholar]
- Zhang W., Yang S. L., Yang M., Herrlinger S., Shao Q., Collar J. L., et al. (2019). “Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors.”. Nat. Commun. 10:2612. 10.1038/s41467-019-10497-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Fan L. X., Li K., Ramchandran R., Calvet J. P., Li X. (2014). “SIRT2 regulates ciliogenesis and contributes to abnormal centrosome amplification caused by loss of polycystin-1.”. Hum. Mol. Genet. 23 1644–1655. 10.1093/hmg/ddt556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg D., Debbache J., Pena-Hernandez R., Antunes A. T., Schaefer S. M., Cheng P. F., et al. (2018). “EZH2-Mediated Primary Cilium Deconstruction Drives Metastatic Melanoma Formation.”. Cancer Cell 34 69–84e14. 10.1016/j.ccell.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Zombor M., Kalmar T., Nagy N., Berenyi M., Telcs B., Maroti Z., et al. (2019). “A novel WDR62 missense mutation in microcephaly with abnormal cortical architecture and review of the literature.”. J. Appl. Genet. 60 151–162. 10.1007/s13353-019-00486-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.