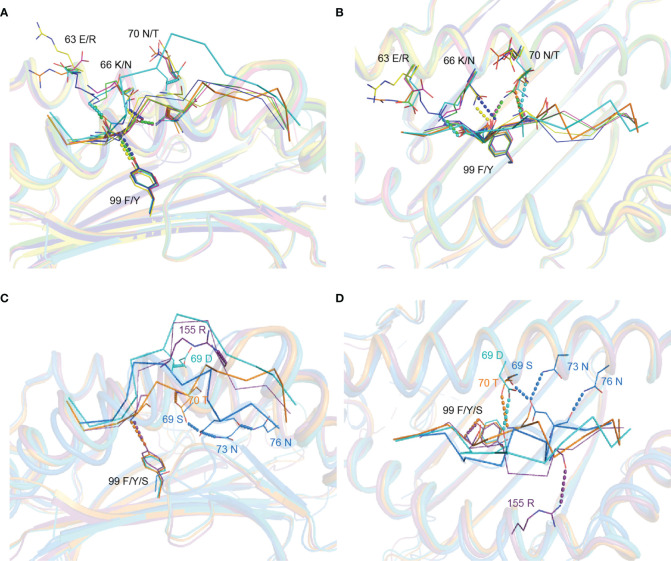
Figure 5.
Analysis of the effect of variant residues between SLA-1*13:01 and SLA-1*04:01 on MHC-I binding peptides. (A, B) Structural comparison between determined pSLA-1*13:01RW12 (cyan), pSLA-1*04:01RW12 (orange, PDB code 6LF8), pSLA-1*04:01MY9 (blue, PDB code 6KWK), pSLA-1*13:01(F99Y)NW9 (green, PDB code 6KWN) and pSLA-1*13:01EW9 (magenta, PDB code 6KWO). The dotted lines of different colors indicate the interaction between the peptides and the residues of the antigen binding groove in different structures. (C, D) Structural comparison between determined pSLA-1*13:01RW12 (cyan), pSLA-1*04:01RW12 (orange, PDB code 6LF8), pMamu*B17IW11 (purple, PDB code 3RWD) and pBF2*21:01GL11 (purple blue, PDB code 2YF1). The dotted lines of different colors indicate the interaction between the peptides and the residues of the antigen binding groove in different structures.