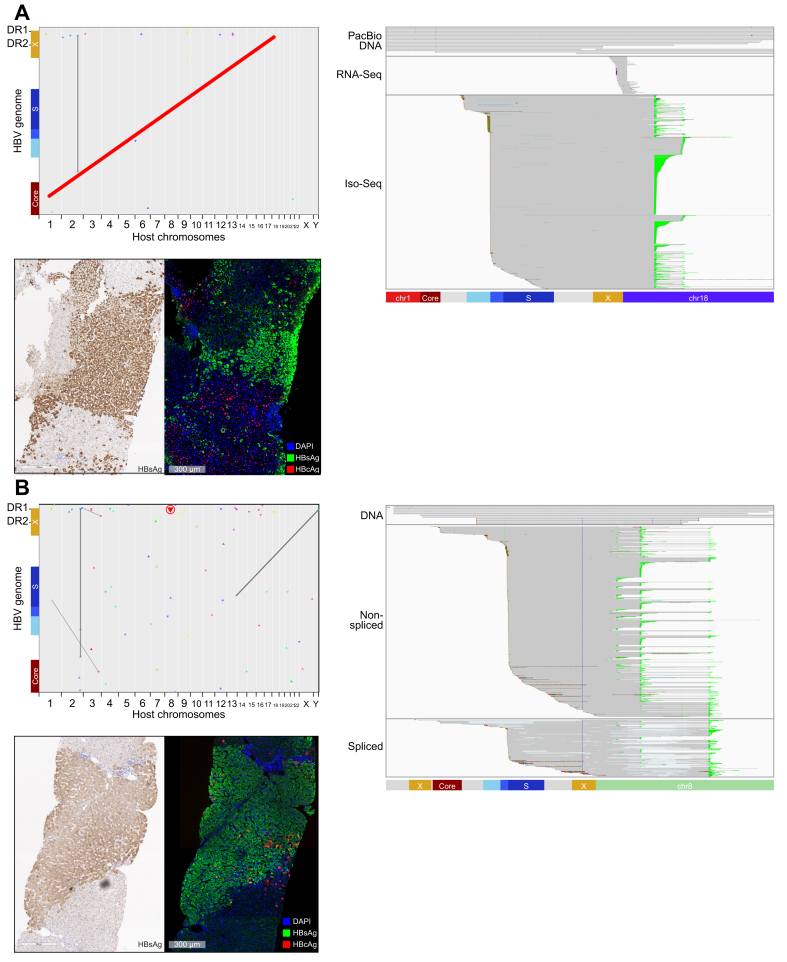
Fig. 5.
Identification of a clonally expanded and transcriptionally active HBV-associated chromosomal translocation.
(A) Matching FFPE and fresh frozen baseline biopsies were analysed from a 36-year-old White male who was HBeAg-negative. This patient had HBV DNA load of 7.22 log10 IU/ml and HBsAg load of 3.90 log10 IU/ml. We obtained dozens of unique reads confirming an HBV-associated chromosomal translocation between chromosome 1 and chromosome 18. We also obtained targeted Iso-Seq and bulk RNA-Seq reads indicating transcriptional activity from the same integration. Two FFPE slides were stained. As part of a multiplex panel, HBcAg, HBsAg, and NaKATPase were overlaid. In addition, a chromogenic HBsAg stain was performed. (B) Matching FFPE and fresh frozen baseline biopsies were analysed from a 24-year-old Asian male who was HBeAg-positive. This patient had a HBV DNA load of 7.54 log10 IU/ml and a HBsAg load of 3.74 log10 IU/ml. We obtained a number of reads confirming an integration consisting of 4,119 bp of continuous HBV sequence. We also obtained targeted Iso-Seq reads indicating transcriptional activity from the same integration. Two FFPE slides were stained. As part of a multiplex panel, HBcAg, HBsAg, and NaKATPase were overlaid. In addition, a chromogenic HBsAg stain was performed. FFPE, formalin-fixed paraffin-embedded; RNA-Seq, RNA sequencing; targeted Iso-Seq, targeted long-read RNA-sequencing.