Figure 1. Bulk FRET measurements demonstrate modest contribution by DOT1L-nucleosome acidic patch interaction.
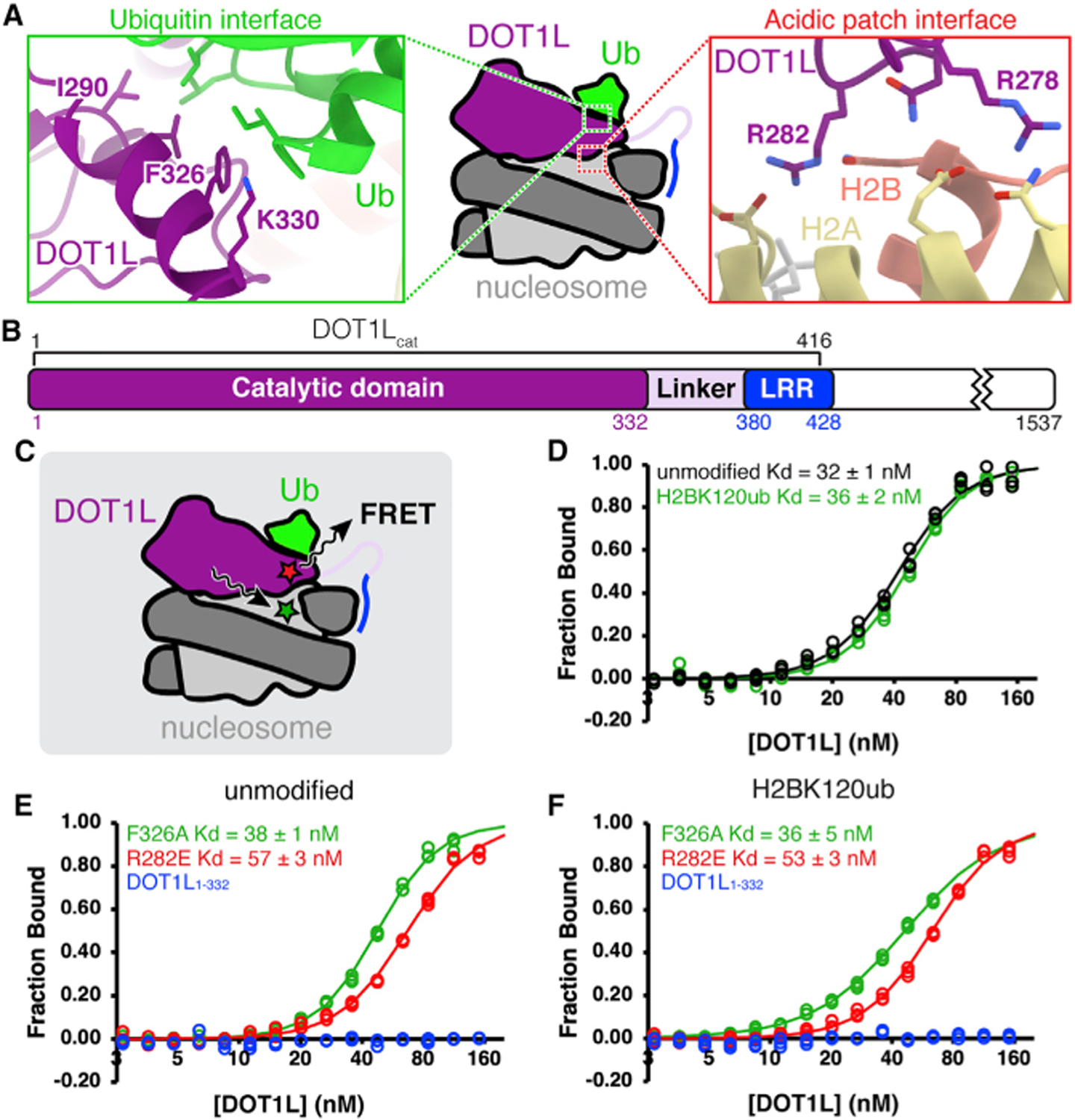
(A) DOT1L-H2BK120ub nucleosome interaction interfaces (PDB: 6NN6). Ub = ubiquitin.
(B) Protein domain map of human DOT1L, with DOT1Lcat region used for binding assays denoted. LRR = lysine-rich region.
(C) Experimental setup for bulk FRET binding assays. FRET between Alexa Fluor 488-labeled nucleosomes (green star) and Atto647N-labeled DOT1L (red star) used to quantitate DOT1L-nucleosome binding affinity.
(D) Normalized binding curves and fits for wild-type DOT1Lcat binding to unmodified or H2BK120ub nucleosomes.
(E and F) Normalized binding curves and fits for mutants of DOT1Lcat or DOT1L1–332 binding to (E) unmodified or (F) H2BK120ub nucleosomes. All nucleosomes contain an H3K79Nle substitution. Individual data points are shown for three independent titrations.
See also Figure S1.
