Figure 3. DOT1L lysine-rich region, ubiquitin- and acidic patch-binding, and SAM stabilize higher FRET DOT1L-nucleosome conformations in single-molecule studies.
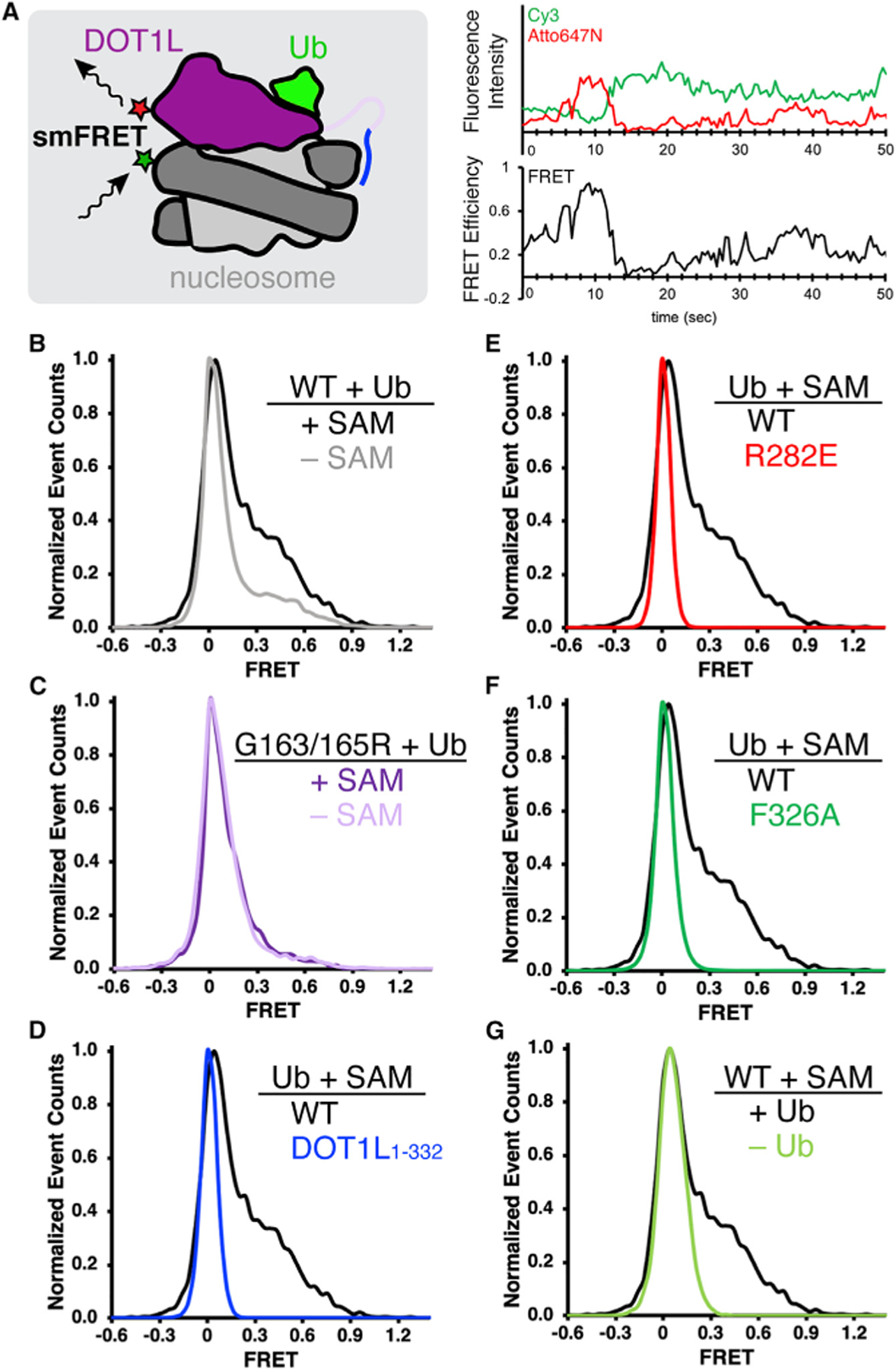
(A) Experimental setup for single-molecule FRET (smFRET) measurements (left), where FRET between Cy3-labeled nucleosomes (green star) and Atto647N-labeled DOT1L (red star) detects DOT1L-nucleosome binding conformation. An example of fluorescence intensity and smFRET time trajectories shows dynamic DOT1L-nucleosome binding. Ub = ubiquitin.
(B–F) (B and C) Normalized histograms comparing DOT1L-H2BK120ub nucleosome interactions with and without SAM, using (B) wild-type (WT) DOT1Lcat (n = 161 and 141 smFRET time trajectories for with and without SAM, respectively) or (C) SAM-binding deficient G163/165R DOT1L (n = 123 and 88 for with and without SAM, respectively), or (D–F) Normalized histograms comparing wild-type DOT1L-H2BK120ub interactions in the presence of SAM with (D) DOT1L1–332 (n = 868), (E) DOT1L R282E acidic patch interface mutant (n = 3,348), or (F) DOT1L ubiquitin interface F326A mutant (n = 2,882).
(G) Normalized histogram comparing wild-type DOT1L binding to H2BK120ub or unmodified nucleosomes (n = 1,251) in the presence of SAM.
See also Figures S1 and S3.
