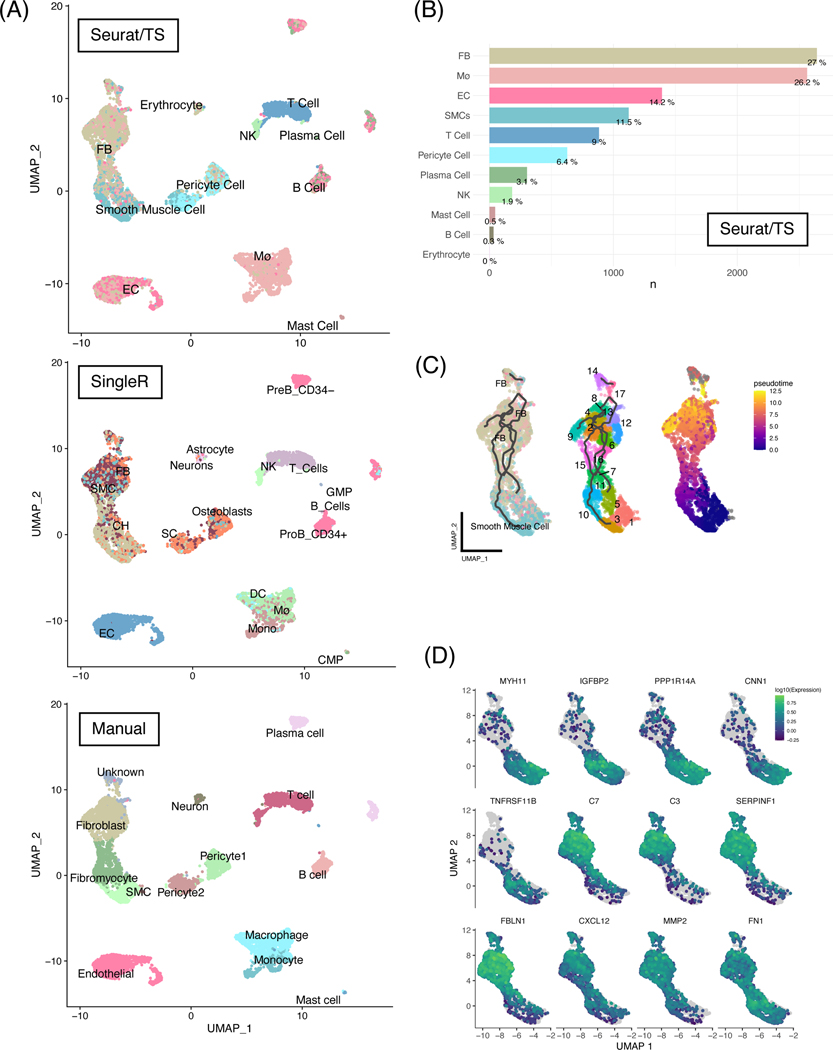
Figure 1.
Unbiased automatic cell labeling of human coronary scRNA data using ‘Seurat v4’ had high concordance with manual labels and pseudotemporal and ultra-fine clustering reveals FB-like cells derivation from SMCs.
(A) Top panel: UMAP clustering of 9798 cells derived from human coronary artery explants with labels based on Seurat v4 using the Tabula sapiens reference; middle panel: automatic labels provided by singleR using Human Primary Cell Atlas; bottom: cluster-based manual annotation provided by original authors Wirka et al. (B) Population breakdown by percentage based on Seurat/TS labels. (C) Left panel: RNA trajectory (line) shows direct paths from the SMCs toward FBs; middle: ultra-fine clustering shows the logical transition stages (microclusters) from SMCs to FBs, and each cluster is numbered for clarity; right: pseudotemporal analysis confirms that the cell and clusters existing along a logical single-cell continuum. (D) Selected genes that were shown to vary over pseudotime by Moran’s I test were visualized. Abbreviations: SMC: smooth muscle cells, EC: endothelial cells, FB: fibroblasts, Mø: macrophages.