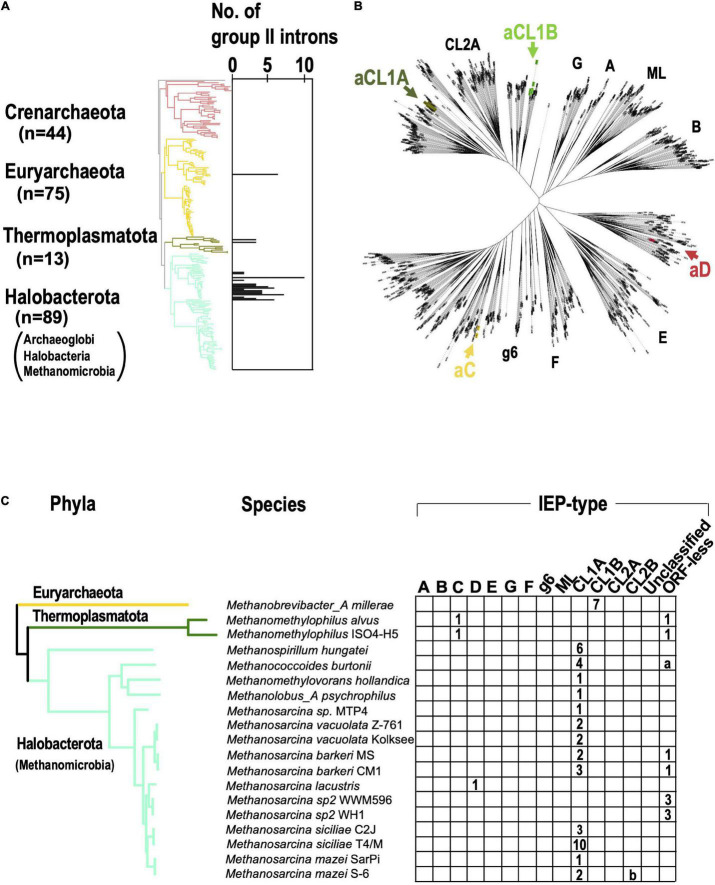
FIGURE 4.
G2Is are increased in certain archaeal phyla. (A) Increases in numbers of G2Is in specific archaeal phyla. The numbers of G2Is in complete archaeal genomes (222 genomes) are shown. Archaeal phyla are shown on the left and each corresponding branch on the archaeal phylogenic tree is colored. The numbers in brackets represent the number of genomes in each phylum. (B) Positions of archaeal IEPs on an unrooted phylogenetic tree of the representative IEP sets (see Figure 2A in details). aCL1A (archaeal CL1A), aCL1B (archaeal CL1B), aC (archaeal-C), and aD (archaeal-D). (C) Distribution of types of IEPs in the archaeal phylogeny. The distribution of the types of IEPs in 19 archaeal genomes that contain G2I(s) is shown. Numbers of G2I(s) per IEP type are also shown in each box. a: Analysis of the Methanococcoides burtonii genome with our pipeline incorrectly detected four ORF-less G2Is. These were parts of CL1A-type G2Is. b: Analysis of the Methanosarcina mazei S-6 genome with our pipeline incorrectly detected one G2I classified as the CL2B type, because the G2I was divided by a transposase. A detailed analysis revealed that it was a CL1A-type G2I.