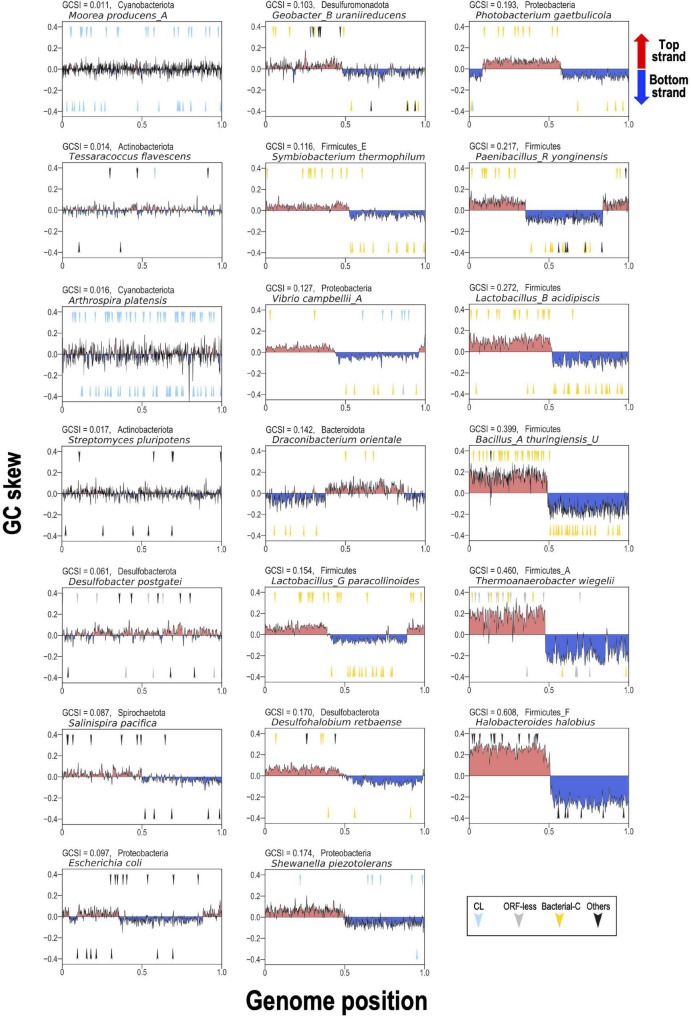
FIGURE 7.
GC skew and insertion positions of G2Is in 20 representative bacterial genomes. In the boxes, the vertical axis shows the GC skew index of each genome, and the boxes are arranged in ascending order from the upper left according to the GCSI. The horizontal axis shows the relative position of each genome; the start position of the base sequence in each GenBank file is set to 0, and the end position is set to 1. The arrow in the upper half of each plot represents the insertion of G2Is into the top strand, and the arrow in the lower half represents the insertion of G2Is into the bottom strand. The colors of the arrows and the classification of G2Is are as follows: CL type (blue), ORF-less (gray), bacterial-C type (yellow), and others (black). The GCSI, bacterial phylogeny, and species name are shown in the upper left of each box. The RefSeq genome accession numbers are as follows: Moorea producens_A: NZ_CP017599.1; Tessaracoccus flavescens: NZ_CP019607.1; Arthrospira platensis: NC_016640.1; Streptomyces pluripotens: NZ_CP021080.1; Desulfobacter postgatei: NZ_CM001488.1; Salinispira pacifica: NC_023035.1; Escherichia coli: NC_011750.1; Geobacter_B uraniireducens: NC_009483.1; Symbiobacterium thermophilum: NC_006177.1; Vibrio campbellii_A: NC_009784.1; Draconibacterium orientale: NZ_CP007451.1; Lactobacillus_G paracollinoides: NZ_CP014915.1; Desulfohalobium retbaense: NC_013223.1; Shewanella piezotolerans: NC_011566.1; Photobacterium gaetbulicola: NZ_CP005974.1; Paenibacillus_R yonginensis: NZ_CP014167.1; Lactobacillus_B acidipiscis: NZ_LT630287.1; Bacillus_A thuringiensis_U: NC_022873.1; Thermoanaerobacter wiegelii: NC_015958.1; Halobacteroides halobius: NC_019978.1.