Figure 4. Knocking out baiH in gnotobiotic mice.
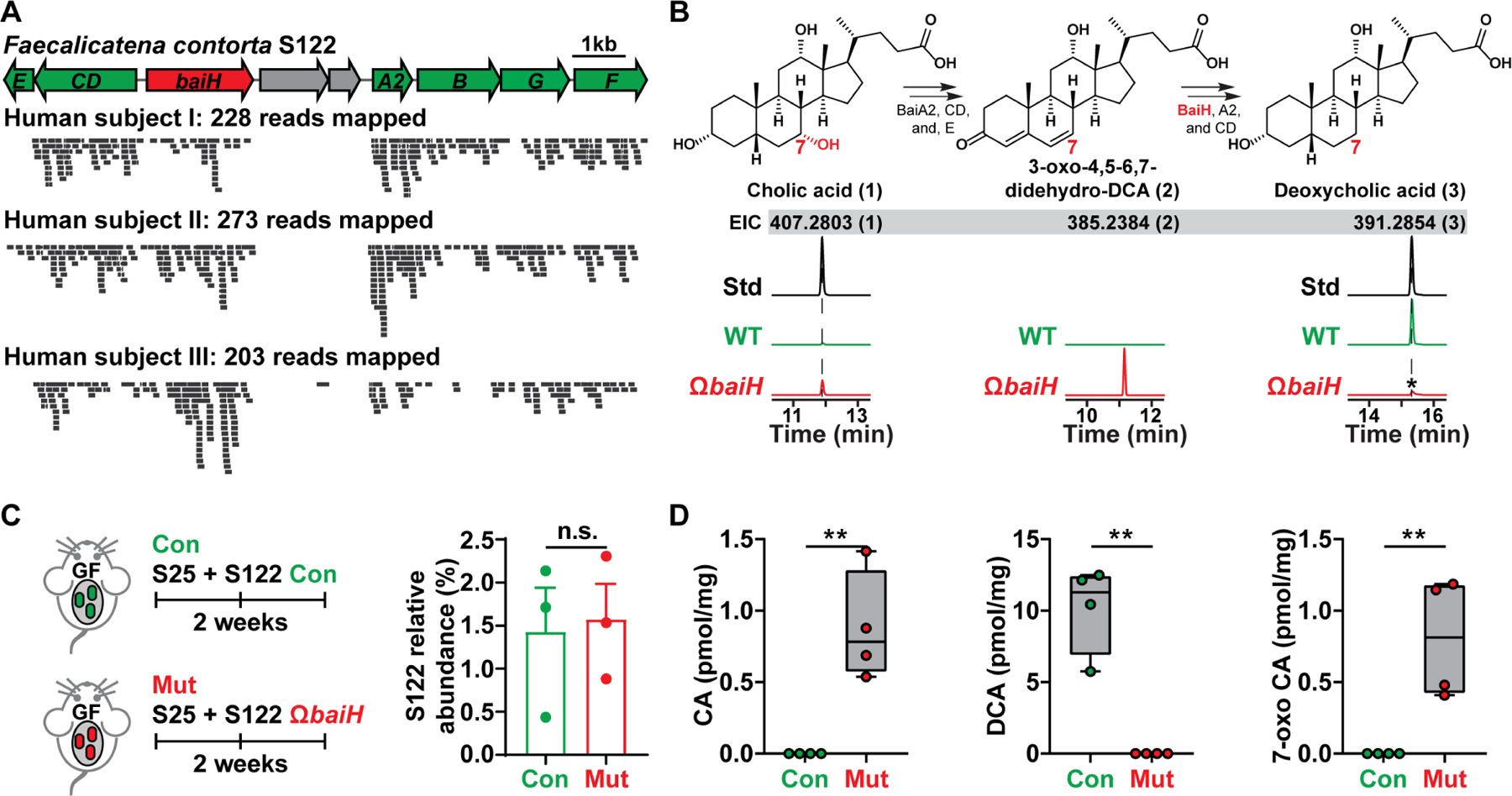
(A) The orientation of the S122 bai operon for bile acid 7α-dehydroxylation. The mutated gene baiH (by Group II intron) is highlighted in red. The S122 bai operon is actively transcribed under host colonization, and three representative results of metatranscriptomic analyses of the S122 bai operon are shown.
(B) The biosynthetic scheme of bile acid 7α-dehydroxylation. The baiH encodes an oxidoreductase that reduces the 6,7-olefinic bond of the intermediate 3-oxo-4,5–6,7-didehydro-DCA (2, EIC: 385.2384). The S122 ΩbaiH mutant accumulates the predicted intermediate (2, EIC: 385.2384) and no longer converts CA (1, EIC: 407.2803) to DCA (3, EIC: 391.2854) in vitro. The structure of the intermediate (2) was determined by comparing its retention time and exact mass to the published literature. The asterisk indicates a residual amount of DCA that is a contaminant from the CA chemical standard. EIC: extracted ion chromatogram.
(C) Germ-free C57BL/6J mice (n = 3 or 4 per group) were co-colonized with S25 plus the S122 control (Con) or ΩbaiH mutant (Mut) (by Group II intron) strain. The relative abundances of S122 in the control and mutant group were assessed by 16S rRNA sequencing and were comparable.
(D) Depleting baiH using Group II intron abolishes gut 7α-dehydroxylating activity and modifies gut bile acid pool in gnotobiotic mice. CA, DCA, and 7-oxo CA (see Data S1E for their structures) were quantified using LCMS.
Data in (C) and (D) were analyzed using unpaired two-tailed Student’s T-test. The asterisk indicates p-value < 0.05 (*) or < 0.01 (**). The numbering of the strains corresponds to the strain information shown in Table S1.
