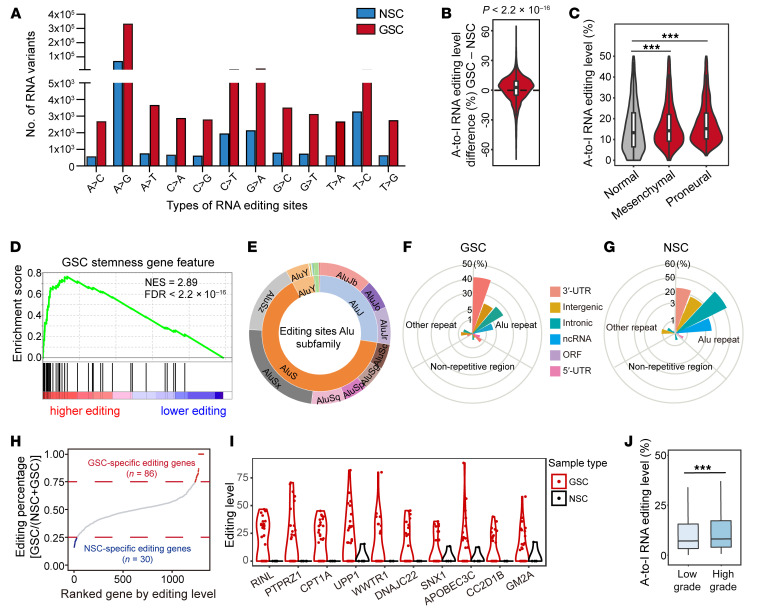
Figure 1. Global landscapes of A-to-I RNA editing in GSCs.
See also Supplemental Figure 1. (A) Distribution of 12 types of RNA editing events in GBM stem cells (GSCs) and neural stem cells (NSCs). Data represent the mean number of inferred RNA editing events detected in GSCs and NSCs. (B) The comparative distribution of A-to-I RNA editing levels in GSCs relative to NSCs. P value was determined by Wilcoxon’s signed-rank test. (C) The distribution of A-to-I RNA editing levels in normal NSCs and different subtypes of GSCs. Statistical significance was determined by Wilcoxon’s signed-rank test and adjusted using the Bonferroni method. ***P < 0.001. (D) Gene set enrichment analysis of GSC stemness gene feature with different A-to-I RNA editing patterns. (E) The distribution of A-to-I RNA editing events in different types of Alu subfamilies. (F) The distribution and percentage of A-to-I RNA editing events in different regions of RNA transcripts in GSCs. (G) The distribution and percentage of A-to-I RNA editing events in different regions on RNA transcripts in NSCs. (H) The distribution of editing percentage of genes ranked by editing level. Eighty-six genes were defined as having GSC-specific editing, and 30 genes were defined as having NSC-specific editing. (I) The 10 genes, among those defined in H as having GSC-specific editing, with the highest levels of editing and dysregulation in cancer biology are depicted. (J) Relative frequency of A-to-I RNA editing events in low-grade and high-grade gliomas in TCGA (17). Statistical significance was determined by Wilcoxon’s signed-rank test. ***P < 0.001.