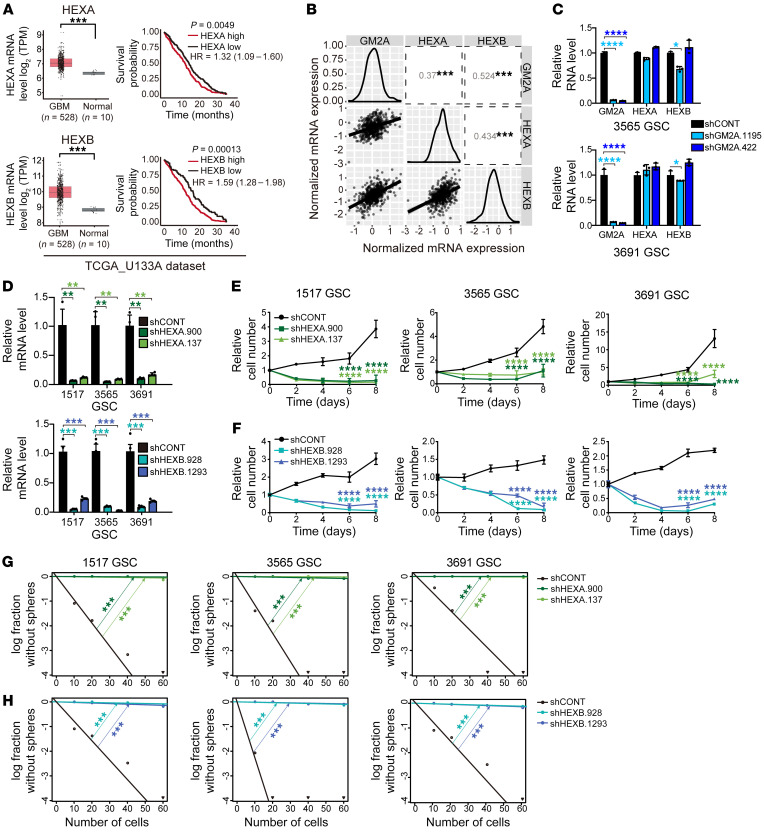
Figure 5. The GM2 ganglioside catabolic pathway maintains GSCs.
(A) Left: Gene expression levels of HEXA and HEXB in normal brain and gliomas in TCGA. Statistical significance was determined by Wilcoxon’s signed-rank test. ***P < 0.01. Right: Kaplan-Meier survival curves of glioma patients with high or low HEXA or HEXB expression in TCGA. P value was determined by log-rank test. (B) Pairwise correlation analysis between GM2A, HEXA, and HEXB gene expression data from TCGA GBM data sets. Correlation coefficient (R) values are shown. (C) mRNA expression of HEXA and HEXB in GSCs (3565 and 3691) transduced with shCONT or shGM2A. Quantitative data from 3 independent experiments are shown as mean ± SD (error bars). n = 3. Statistical significance was determined by ANOVA. *P < 0.05, ****P < 0.0001. (D) HEXA (top) and HEXB (bottom) mRNA expression in GSCs (1517, 3565, and 3691) transduced with shCONT, shHEXA, or shHEXB. n = 4. Quantitative data from 4 independent experiments are shown as mean ± SD (error bars). Statistical significance was determined by ANOVA. **P < 0.01, ***P < 0.001. (E) Proliferation of GSCs (1517, 3565, and 3691) transduced with shCONT or shHEXA determined by CellTiter-Glo. n = 5. Quantitative data from 5 technical experiments are shown as mean ± SD (error bars). Statistical significance was determined by 2-way ANOVA with Dunnett’s multiple-comparison test. ****P < 0.0001. (F) Proliferation of GSCs (1517, 3565, and 3691) transduced with shCONT or shHEXB determined by CellTiter-Glo. n = 5. Quantitative data from 5 technical experiments are shown as mean ± SD (error bars). Statistical significance was determined by 2-way ANOVA with Dunnett’s multiple-comparison test. ****P < 0.0001. (G) ELDA for in vitro sphere formation of GSCs (1517, 3565, and 3691) transduced with shCONT or shHEXA. n = 24. Pairwise tests for differences in stem cell frequencies. ***P < 0.001. (H) Analyses identical to those in G were performed for HEXB.