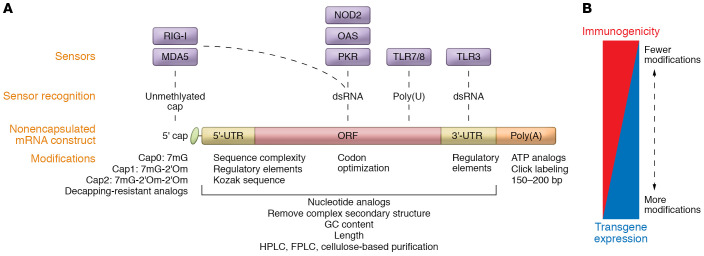
Figure 1. Modifications of mRNA vaccines for enhanced antitumor immunity.
Several strategies have been used to chemically modify mRNA constructs to optimally balance antigen expression with innate immune recognition of the mRNA construct itself. (A) Manipulations that enhance antigen expression include modification in the 5′ cap through anti-reverse cap analogs, methylation of start nucleotides, and decapping-resistant analogs; modification in untranslated regions through the removal of long stem-loop-like structures with high GC content, insertion of an internal ribosomal entry site within the 5′-UTR, and inclusion of a Kozak sequence upstream of the start codon; modification in the open reading frame (ORF) through codon optimization based on target cell tRNA abundance; and modification of the poly(A) tail by incorporation of ATP analogs and click-labeling with fluorescent dyes. The ideal polyA length within human cells is approximately 120 bases. Use of nucleotide analogs, sequence complexity, GC content, and length modifications can reduce detection by innate sensors such as TLR7/8 or TLR3 or cytoplasmic sensors such as RIG-I, MDA5, OAS, NOD2, and PKR. Purification of in vitro–transcribed RNAs by HPLC, FPLC, or cellulose-based methods can further remove contaminating dsRNA products that would engage these sensors. (B) While these alterations result in greater transgene expression and less immunogenicity to the mRNA construct, the optimal combination of modifications and balance of the two to yield a therapeutic advantage is still an open question as it relates to optimization of cancer vaccines.