FIGURE 4.
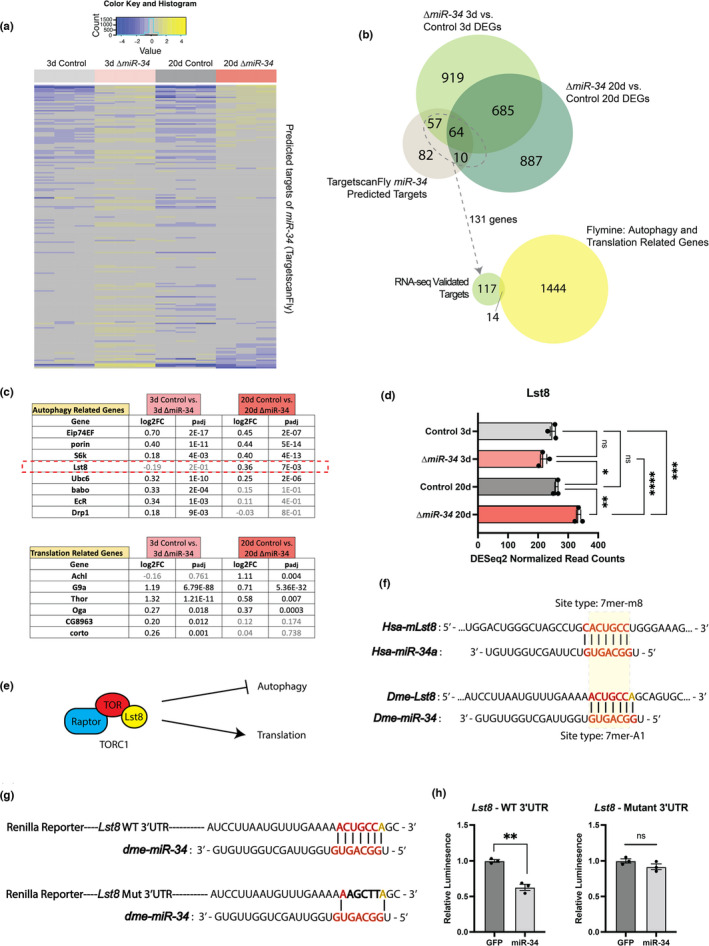
miR‐34 is predicted to target TORC1 subunit, Lst8. (a) Heat map comparing transcript levels for all TargetscanFly predicted targets of Drosophila miR‐34 (b) Venn Diagram (top) presenting the overlap between genes significantly upregulated in the 3d (miR‐34 vs. control) and 20d (miR‐34 vs. control) brain vs. predicted targets of Drosophila miR‐34. Venn Diagram (bottom) presenting overlap between RNA‐seq verified targets of miR‐34 and Translation‐related and Autophagy‐related gene lists derived from Flymine database. (c) RNA‐seq validated targets of miR‐34 associated with autophagy and translation based on curated gene lists from the Flymine database. (d) RNA‐seq normalized reads for Lst8. Lst8 is significantly upregulated in the 20d miR‐34 mutant brain (n = 3 biological replicates, mean ±SEM, two‐way ANOVA (Age: F(1,8) = 60.88, p < 0.0001 | Genotype: F(1,8) = 6.968, p = 0.0297 | Interaction: F(1,8) = 40.04, p 0.0002) with Tukey's multiple comparison test). (e) Schematic showing the role of TORC1 in regulating both translation and autophagy. (f) 3’UTR Lst8 and mLst8 seed sequences for miR‐34. (g) Wild‐type and mutant seed sequences for Lst8 3’UTR reporter constructs. (h) Luciferase reporter assay for WT and miR‐34 mutant 3’UTR with co‐expression of miR‐34 or GFP control. Upregulation of miR‐34 significantly reduced the activity of Renilla luciferase fused to only the WT 3’UTR of Lst8 (n=3 wells/condition, mean ± SEM, Student's t test). Significance: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
