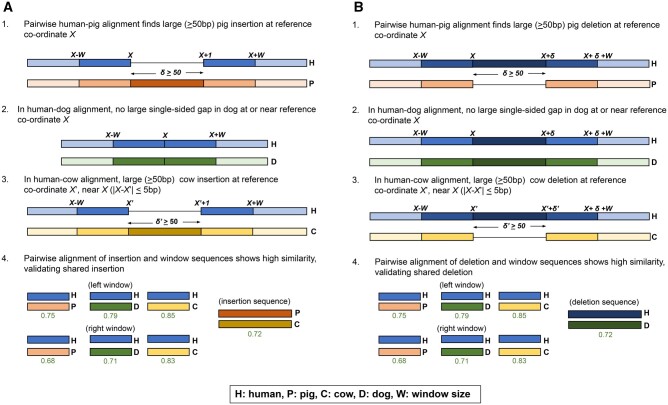
Fig. 5.
Champagne’s indel verification method. (A) Shared insertion between pig and cow detected by Champagne that is absent in dog. (1) We first identify the presence of this insertion by finding a single-sided human gap in the human–pig orthologous chains, at human coordinate X. (2) Next, we find that there is no such single-sided gap in dog chain near X, we mark the insertion as likely absent in dog. (3) Next, we navigate to coordinate X in the human–cow chains, and check for a large (similar-sized) gap at , within a 5-bp range of X. Finding such a gap, indicating an insertion, we mark the insertion as likely present in cow. (4) Finally, we perform a direct sequence comparison for sequence similarity. We extract a 30-bp-sized “window” sequence from either side of the insertion coordinate X in human, either side of the corresponding insertion coordinate in dog, and either side of the insertion itself in cow and pig. We also extract the sequence of the insertion itself in cow and pig. We then align the reference window sequences against each other species’ window sequences. Similarly, we align pig’s insertion sequence against cow’s insertion sequence. For each species in which we marked the indel as present, if the minimum sequence similarity for the left window, right window, and insertion (if the insertion is present) is greater than our stipulated threshold, we mark the species as definitively “+.” For each species in which we marked the indel as absent, if the sequence similarities for the left window and right window are greater than our stipulated threshold, we mark the species as definitively “−.” In either case, if a comparison fails to meet the threshold, we mark the species as “?.” (B) Symmetrical process for finding shared deletions.