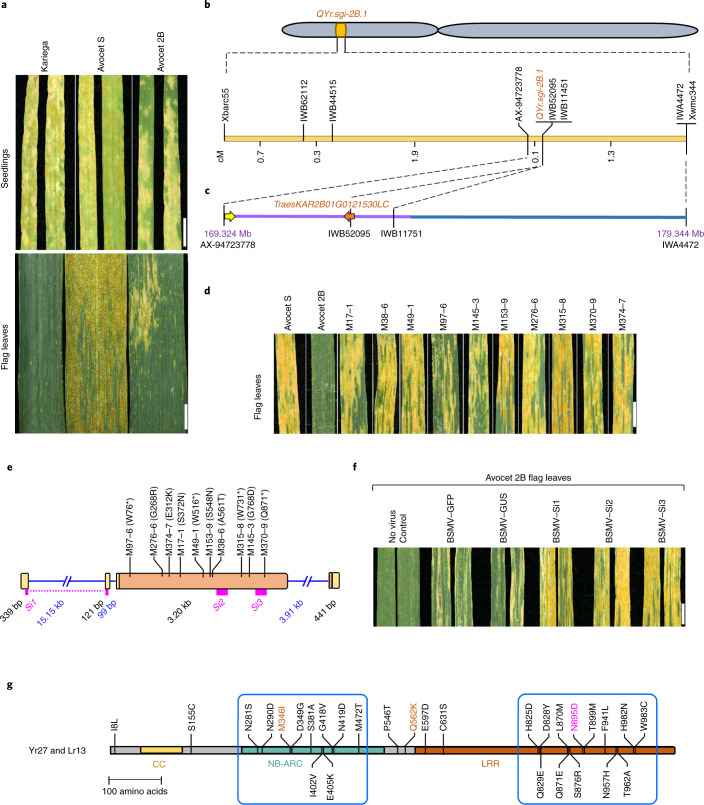
Fig. 1. Assembly-guided cloning of QYr.sgi-2B.1.
a, Stripe rust symptoms on Kariega, Avocet S and Avocet 2B in seedlings (upper panel) and adult plants (lower panel). Scale bars, 1 cm. b, Genetic map of the QYr.sgi-2B.1 region. Numbers between markers indicate genetic distances. c, Physical interval of the QYr.sgi-2B.1 region. Numbers indicate positions (in megabases) in the Kariega 2B pseudomolecule. The interval contained 93 candidate genes that were determined based on the annotated high-confidence gene models (92 high-confidence genes) and the NLR annotation. The NLR annotation identified one additional gene (TraesKAR2B01G0121530LC) that was annotated as a low-confidence gene. The two arrows indicate NLR-encoding genes that were prioritized for gene validation. Purple and blue bars indicate two primary HiFi contigs (47.7 Mb and 19.5 Mb, respectively) that spanned the target interval. d, Stripe rust inoculation on Avocet 2B and ten independent EMS mutants that lost the QYr.sgi-2B.1-mediated resistance. Scale bar, 1 cm. e, Gene structure of QYr.sgi-2B.1. Boxes, exons; lines, introns; yellow boxes, 5′ UTR and 3′ UTR; orange boxes, coding sequence. Positions of the corresponding amino acid changes in the ten loss-of-function mutants are indicated. Si1, Si2 and Si3 denote three VIGS constructs. f, Representative images showing the results of the VIGS experiment. The control ‘no virus’ contained no BSMV. BSMV–GFP and BSMV–GUS denote controls with a GFP or GUS silencing construct, respectively. BSMV–Si1, BSMV–Si2 and BSMV–Si3 denote three different silencing constructs that target QYr.sgi-2B.1. The specificity of Si1–Si3 was evaluated using the Kariega assembly. The image shows flag leaves 15 days after inoculation with an avirulent Pst isolate. Chlorotic areas on the GUS and GFP controls represent virus symptoms. Scale bar, 1 cm. g, Comparison of the Yr27 and Lr13 protein sequences. The first residue is the one present in Yr27, and the second residue is the one present in Lr13. Blue boxes indicate regions with increased polymorphism density. The two amino acids shown in brown have been found to determine the Lr13-specificity2. The residue shown in magenta is found only in Yr27.