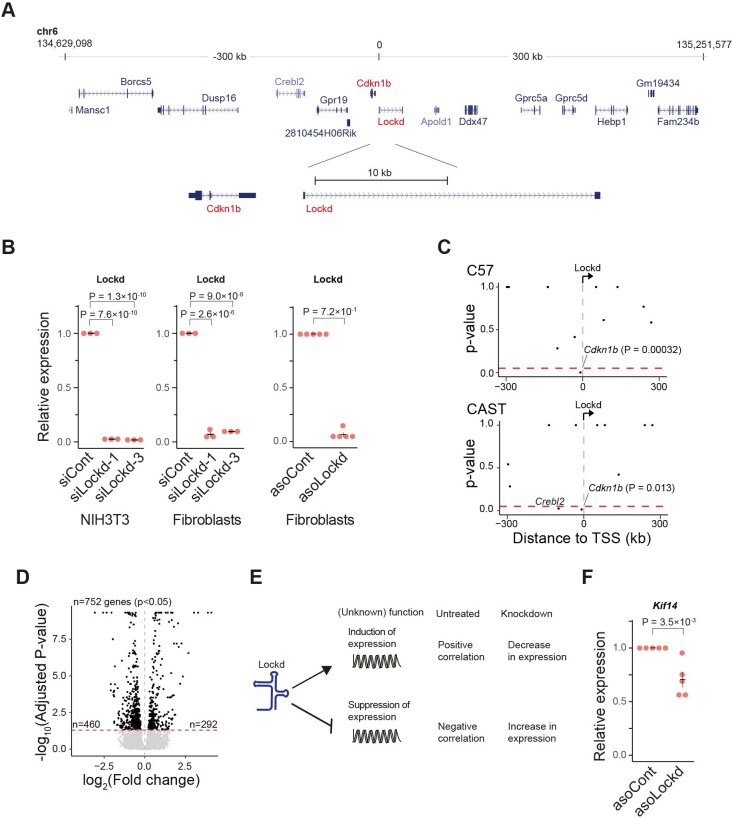
Extended Data Fig. 6. Analysis of the Lockd harboring genomic loci.
(A) A schematic from the UCSC genome browser representing the Cdkn1b-Lockd gene locus. (B) Relative expression of Lockd upon; siRNA induced knockdown in NIH/3T3 cells (n = 3 biologically independent samples), primary fibroblasts (n = 3 biologically independent samples) and ASO induced knockdown in primary fibroblasts (n = 5 biologically independent samples) measured by RT-qPCR. (C) Scatter plot showing p-values for co-expression of Lockd against genes in proximity for the CAST and C57 genomes (Fisher’s exact test, ± 300 kb of TSS-Lockd, genes filtered for > = 3 allelic counts). The red dashed lines denote threshold for significance (p < 0.05). (D) Scatter plot showing the magnitude (x-axis, SCDE maximum likelihood estimate of the fold change) against significance levels (y-axis, SCDE p-value, two-sided test using the multiple testing corrected (Holm procedure) z-score) for shLockd contrasted shControl treated NIH/3T3 cells. The red dashed line denotes threshold for significance (p < 0.05). Black and grey colored genes represent significant and non-significant fold changes, respectively. (E) Schematic representation for the intersection of fold change (shLockd / shControl) against gene-gene correlations (in shControl treated NIH/3T3 cells). (F) Relative expression of Kif14 upon ASO induced knockdown of Lockd, measured by RT-qPCR (n = 5 biologically independent samples). B, F; data presented as mean values + /- s.e.m., p-values represent a two-sided Student’s t-test.