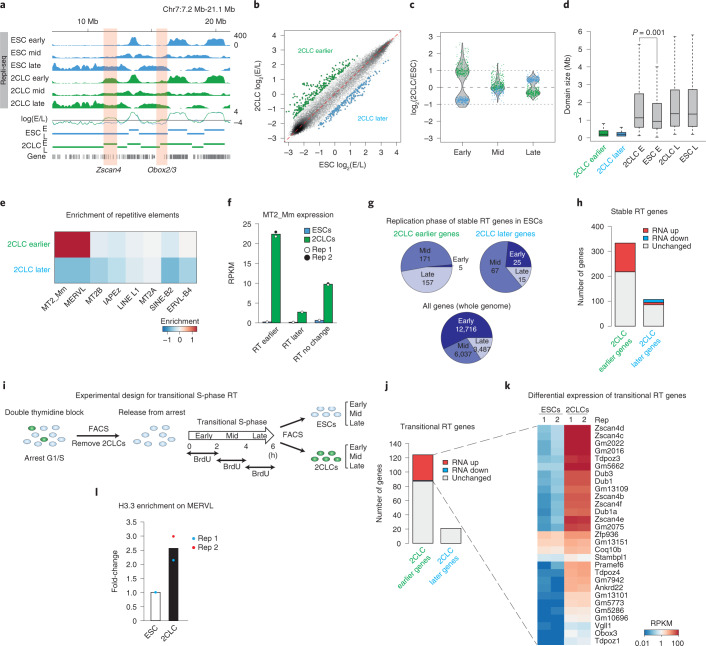
Fig. 4. 2CLCs display changes in RT and slowing replication fork speed promotes reprogramming to totipotency during SCNT.
a, Repli-seq tracks indicating early:late ratio as log2(E/L) for ESCs and 2CLCs. Horizontal lines indicate early (E) and late (L) replicating domains. Orange highlights regions of differential RT. b, Comparison of log2(E/L) between 2CLCs and ESCs at 100-kb bins across the genome. Green and blue points are regions of differential RT (twofold cutoff). c, Violin plot of log(fold differences) between 2CLSs and ESCs at early, mid and late S-phase over twofold differential log2(E/L) 100-kb bins. d, Early replicating domains are larger in 2CLCs. Boxplot of domain sizes for differential RT shows early or late domains from two biological replicates. Boxes show IQR between first and third quartiles, horizontal line shows the median and whiskers show Q3 + 1.5 × IQR and Q1 − 1.5 × IQR. Statistical significance comparing domain sizes was by two-sided Student’s t-test. e, Genomic regions replicating earlier in 2CLCs are enriched in MT2_Mm and MERVL. Heatmap showing log2(fold enrichment) of repeats within earlier and later RT regions. f, Barplot of average expression (RPKM (reads per kilobase per million mapped reads)) in ESCs and 2CLCs of MT2_Mm repeats, the replication of which showed earlier, later or no shift in 2CLCs. Points indicate values for biological RNA-seq replicates. g, Shift of most genes to earlier replication in 2CLCs replicating in mid or late S-phase in ESCs. Proportions are shown by the numbers of genes according to their replication profile in ESCs, for the gene sets that shifted to earlier or later timing in 2CLCs. h, Genes replicating earlier in 2CLCs tend to be upregulated in 2CLCs. The barplot shows number of genes shifting to earlier or later RT in 2CLCs according to their expression changes (115 upregulated, 0 downregulated and 218 unchanged). i, Strategy to map replication timing in emerging 2CLCs during their transitional S-phase. j, Barplot depicting number of genes shifting to earlier or later RT during the transitional S-phase, during which 2CLCs emerge, according to their expression changes (29 upregulated, 0 downregulated and 95 unchanged) in 2CLCs. k, Most upregulated genes showing replication shift to earlier in transitional S-phase are repressed in ESCs. Heatmap depicts differential gene expression of genes shifting to earlier RT during the transitional S-phase. RPKM derive from two biological replicates. l, Relative H3.3 enrichment in ESCs and 2CLCs expressing SNAP-tagged-H3.3 analyzed by qPCR CUT&RUN. Dots represent biological replicates.