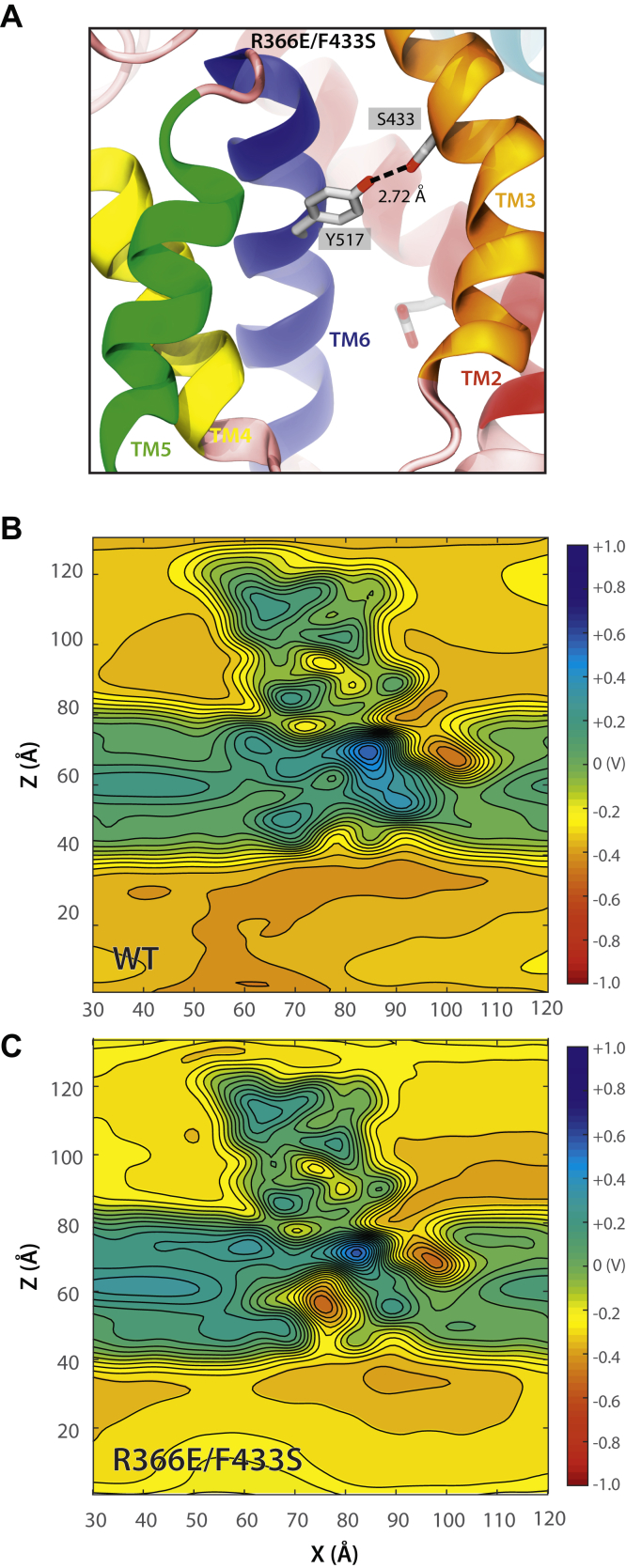
Figure 6.
The Escherichia coli YidC R366E/F433S protein with a negative charge in its groove is functional.A, snapshot of the equilibrated Escherichia coli YidC R366E/F433S system simulated in a hydrated POPE:POPG (75:25) bilayer (phosphopholipids and water are not shown; simulation S3b at ∼28 °C in Table 1). An interaction between the side chain of F433S (suppressor mutation) and the tyrosine residue at site 517 is observed during the simulation trajectory (Fig. S6). This interaction might stabilize YidC’s groove. B and C, two-dimensional contour plot of the averaged electrostatic potential computed during 100-ns long simulations of E. coli YidC at ∼28 °C with the protein backbone constrained (WT and R366E/F433S systems in B and C, respectively; simulations S1a and S3a in Table 1). The map corresponds to the electrostatic potential of a slice perpendicular to the membrane plane passing through the center of the protein near site 366. Twenty contour lines are drawn over the range of voltages. All values above or below the scale limits (color scale) are shown at the same level. The electrostatic potential for the R366E/F433S system is negative near the 366 site (C). POPE:POPG, palmitoyloleoyl-phosphatidylethanolamine:palmitoyloleoyl-phosphatidylglycerol.