Abstract
In recent years, generative adversarial networks (GANs) have gained tremendous popularity for various imaging related tasks such as artificial image generation to support AI training. GANs are especially useful for medical imaging–related tasks where training datasets are usually limited in size and heavily imbalanced against the diseased class. We present a systematic review, following the PRISMA guidelines, of recent GAN architectures used for medical image analysis to help the readers in making an informed decision before employing GANs in developing medical image classification and segmentation models. We have extracted 54 papers that highlight the capabilities and application of GANs in medical imaging from January 2015 to August 2020 and inclusion criteria for meta-analysis. Our results show four main architectures of GAN that are used for segmentation or classification in medical imaging. We provide a comprehensive overview of recent trends in the application of GANs in clinical diagnosis through medical image segmentation and classification and ultimately share experiences for task-based GAN implementations.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-021-00556-w.
Keywords: Generative adversarial networks, Medical imaging, Image segmentation, Image classification, Image generation
Introduction
Generative adversarial networks (GANs) have gained tremendous popularity in the field of image processing and generation since their inception in 2014 [1]. GANs have been adapted for complex image processing tasks like image synthesis, data augmentation, semantic segmentation [2], image translation [3], generative image modeling [4], and domain adaptation. GANs have also been adapted for adversarial learning to build robust deep networks that can handle domain shift or bias in training data [5]. Several survey papers have been written to track and analyze the application of GANs for image processing. Wang et al. surveyed the state-of-the-art GAN techniques in 2017 [6], while Kurach et al. focused on various loss functions, regularization, and normalization techniques being used for GAN implementation [7]. Pan et al. [8] evaluated GANs from three main perspectives: (i) high-quality image generation, (ii) diversity of generated images, and (iii) stability of the training process.
While a majority of the GAN literature focuses on general image processing tasks using public datasets like ImageNet [9] and celebA [10], these architectures have also been gaining popularity in the field of medical image processing for segmentation and classification tasks. GANs have been applied across various imaging modalities including radiographs, CT scans, and MRI. A combined literature search in PubMed, Science Direct, and Google Scholar for GAN and MRI, CT, and X-ray produced approximately 5000, 8500, and 2800 results, respectively, in May 2020. GANs have also been used for medical image data synthesis and generation [11–13]. Our search found only a single recently published review paper [14] on the general application of GANs broadly in medicine, without diving deep onto any specific applications.
In this paper, we present a systematic review of the application of GANs for medical image processing. We focused on two main aspects for our review strategy: (i) the final task (i.e., application) for the GAN-based model, focused on classification, segmentation, or generation, and (ii) the modality of the input image, i.e., as radiograph, CT scan, and MRI. We excluded papers outside of medical image processing tasks as there are review papers already published in this area [15, 16]. To the best of our knowledge, there is no systematic review of GANs in medical imaging that summarizes reported performances and discusses the implementation challenges for segmentation and classification tasks. Our goal is to provide an overview of the significant work in this field in a form that is easily digestible for a broad range of readers and highlight some pathways for quick GAN implementation.
The review is structured as follows: “Evidence Acquisition” provides our methodology for selecting the publications for this systematic review. “Evidence Synthesis” provides the results of our survey. Finally, “Discussion” provides a meta-analysis, discusses advanced GAN architectures, and key research challenges in terms of implementation. In supplemental materials, we provided an introduction of the fundamental terminology and algorithms related to GANs.
Evidence Acquisition
This systematic review was conducted based on the PRISMA guidelines [17], specifically taking into account the rationale, objective, and depth of the discussion of the GAN architecture.
Search Strategy
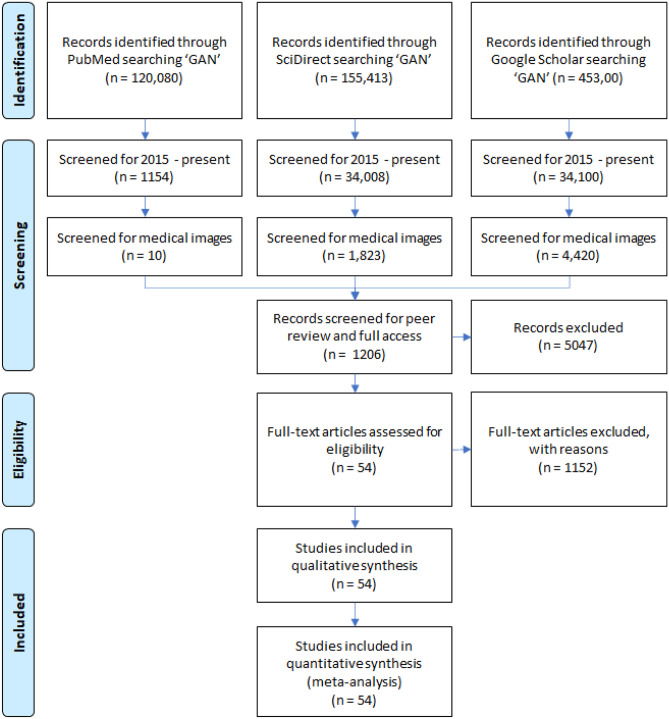
Figure 1 outlines the search strategy. First, the papers for the review were identified from three different sources: PubMed, Science Direct, and Google Scholar. The first screening pass identified papers pertaining to the keywords (“Generative Adversarial Network” or “GAN”) and (“medical image”). The addition of the keyword (“medical image”) was important as it was the specific focus of our review and it also helped to exclude any irrelevant papers from our search with another common explication of (“GAN”): gallium nitride (GaN). Secondly, we excluded papers published before 2015. We used the KEYWORDS and YEAR based on the search criteria in the Publish or Perish software [18] to extract ~ 2000 papers for an internal review for study selection. Publish or Perish is a widely available software that retrieves and analyzes academic papers from various sources like Google Scholar and ScienceDirect. It generates useful analytics of each retrieved citation for a deep review of the literature such as the number of citations per paper and author.
Fig. 1.
PRISMA flow diagram for systematic review of GAN for medical Imaging
Study Selection
Within the internal review for study selection, we selected a total of four reviewers (JJ, AT, TA, and IB). Each reviewer was given a random fourth of the papers for review. We assigned a fifth reviewer (JG) to resolve any conflict between the four. The primary exclusion criteria were (i) papers that were not peer reviewed, such as those in arXiv, and (ii) those that lack methodological details, such as conference abstracts. Finally, the papers eligible to be included in this study were selected using an agreed upon criteria between the raters based on specific targeted tasks image segmentation, classification, or generation. For each paper, we reviewed the GAN model architecture, modifications made to a representative variant of GANs as shown in Table 1, and an application or architecture of interest.
Table 1.
Primary variants of GANs used in medical image analysis
| Representative variants of GANs in medical images | ||
|---|---|---|
| Architecture | Similar variants | Description |
| cGAN (Mirza and Osindero [19]) |
DiagNet (Li et al. [20]) Feature2Mass [13] confidence GAN (Nie and Shen [21]) |
Conditional GANs added conditional information, specifically class label information to the basic GAN architecture allowing the conditional training of GANs and conditional generation of images. This allows the generator to selectively generate an imbalanced class through label input |
| DCGAN (Radford et al. [22]) | Progressive Growing GAN [23] | Deep convolutional GANs explicitly stated the use of convolutional/convolutional-transpose layers and LeakyReLU/ReLU activation layers with batch normalization to stabilize higher resolution GANs training and generation. It has been adopted as a stabilization technique for most current GAN architectures |
| pix2pix [24] and CycleGAN [25] |
Pancreas-GAN [26] ScarGAN [12] SPCGAN (Xing et al. [27]) fixed-point GAN [28] SynSeg-Net (Huo et al. [29]) |
Pix2pix introduced the embedding of whole images as an input to the generator instead of random noise allowing a paired, image-to-image translation. CycleGANs introduced cycle consistency and identity losses allowing for unpaired training of the architecture |
Data Extraction
For benchmarking, the existing GAN approaches, in Tables 2–4, we extracted the following data from each of the selected articles: (a) authors/year of publication, (b) country of author, (c) clinical domain, (d) GAN architecture, (e) application of the task, (f) imaging modality, (g) number of samples for training and testing, and (h) reported performance for papers that generation, classification, and segmentation, respectively. The number of samples reported as the full data-size including training, validation, and testing data. For classification tasks, we extracted area under receiver operating characteristic curve (AUROC) whenever this metric was reported and extracted other metrics such as accuracy, sensitivity, and specificity. When the article contained several experiments, metrics from the experiment with the best performing model were extracted. These items were extracted to enable researchers to quickly find and compare current GAN architectures in their medical field or input modalities of interest.
Table 2.
Benchmarking of the 13 selected manuscripts on GAN generation
| Authors (first/last) | Country | Clinical | GAN | Application | Imaging | Sample |
|---|---|---|---|---|---|---|
| Year | Domain | Architecture | Modality | Sizes | ||
| Kwon/Kim 2019 (Kwon et al. [30]) | South Korea | Neurology | 3D GAN | Whole 3D brain MRI generation | MR | Total: 1421 |
| Train: 1421 | ||||||
| Ossenberg/Grau 2019 (Ossenberg-Engels and Grau [31]) | UK | Cardiology | cGAN | Cardiac MRI translation from end-diastolic to end-systolic | MR | Total: 38,000 |
| Train: 33,500 | ||||||
| Test: 4500 | ||||||
| Chuquicusma/Bagci 2018 (Chuquicusma et al. [11]) | USA | Pulmonology | DCGAN | Synthetic lung nodule generation | CT | Total: 1145 |
| Train: 1145 | ||||||
|
Islam/Zhang 2020 (Islam and Zhang [32]) |
USA | Neurology | DCGAN | Synthetic brain PET image generation | PET | Total: 411 |
| Train: 411 | ||||||
| Lee/Ro 2018 (Lee et al. [13]) | South Korea | Senology | Feature2Mass | Generation of breast masses with target characteristics | MR | Total: 960 |
| Train: 960 | ||||||
| Siddiquee/ Liang 2019 (Siddiquee et al. [28]) | USA | Neurology | fixed-point GAN | Generate new images with fixed points | CT MR | Total 6540 |
| Pulmonology | Train: 4068 | |||||
| Test: 2472 | ||||||
| Yu/Lu 2019 (Yu et al. [33]) | China | Ophthalmology | pix2pix | Multi-input generation of retinal images | Retinal | Total: 141 |
| CycleGAN | Train: 141 | |||||
| Lau/Golden 2018 (Lau et al. [12]) | USA | Cardiology | ScarGAN | Simulation of scar tissue | MR | Total: 159 |
| Train: 159 | ||||||
| Zhang/Liu 2019 (Zhang et al. [34]) | China | Neurology Pulmonology | SkrGAN | Sketch guided image generation | X-ray | Total: 12,709 |
| CT MR | Train: 12,709 | |||||
| Xu/Xu 2019 (Xu et al. [35]) | USA | Pulmonology | 3D multi cGAN | Generation of lung nodules conditioned from the background | CT | Total: 1018 |
| Zhou/Shao 2019 (Zhou et al. [36]) | UAE | Ophthalmology | DR_GAN | Using background and semantic features generate high resolution images with added manipulation | Retinal | |
| Li/Menze 2019 (Li et al. [37]) | Germany | Neurology | DiamondGAN | Multi sequence image synthesis | MRI | Total: 65 |
| Train: 30 | ||||||
| Testing: 35 |
Table 4.
Benchmarking of the 33 selected manuscripts on GAN segmentation
| Authors (first/last) | Country | Clinical | GAN | Application | Imaging | Sample | Performance |
|---|---|---|---|---|---|---|---|
| Year | Domain | Architecture | Modality | Sizes | |||
| Jin/Mollura 2018 (Jin et al. [47]) | USA | Pulmonology | 3D cGAN | Lung nodule generation to augment task | CT | Total: 2000 | Dice: 0.989 |
| Train: 1966 | |||||||
| Test: 34 | |||||||
| Liao/Zhou 2018 (Liao et al. [49]) | USA | Cardiology | 3D GAN | Translation of ICE images into segmentation maps with 2D and 3D information | Ultrasound | Total: 12,196 | Dice: 92.1 |
| Train: 9758 | |||||||
| Test: 2439 | |||||||
| Chaitanya/Konukoglu 2019 (Chaitanya et al. [50]) | Switzerland | Cardiology | cGAN | Cardiac deformation generation to augment task | MR | Total: 50 | Dice: |
| Train: 25–28 | RV:0.844 Myo: 0.825 | ||||||
| Validation: 2 | LV: 0.924 | ||||||
| Test: 20 | |||||||
| Rezaei/Meinel 2017 (Rezaei et al. [51]) | Germany | Neurology | cGAN | Translation of brain tumor images into semantic segmentation maps | MR | Total: 285 | Dice: 0.68 |
| Train: 285 | |||||||
| Gaj/Li 2019 (Gaj et al. [52]) | USA | Rheumatology | cGAN | Translation of knee MRI to segmentation maps | MR | Total: 176 | Dice: 0.88 |
| Train: 122 | |||||||
| Validation: 36 | |||||||
| Test: 18 | |||||||
| Saffari/Puig 2018 (Saffari et al. [53]) | Spain | Senology | cGAN | Translation of breast density images to segmentation maps | X-ray | Total: 410 | Recall: 0.95 |
| Train: 250 | Precision: 0.92 | ||||||
| Validation: 60 | F-score: 0.93 | ||||||
| Test: 100 | |||||||
| Shen/Chen 2019 (Shen et al. [40]) | China | Senology | cGAN | Breast mass generation to augment task | X-ray | Total: 112 | Sen: 0.9619 |
| Train: 63 | Spe: 0.9637 | ||||||
| Test: 49 | Dice: 0.9628 | ||||||
| Nie/Shen 2020 (Nie and Shen [21]) | USA | Urology | confidence GAN | Confidence map generation for segmentation | MR | Total: 404 | Dice: 0.8952 |
| Train: 240 | |||||||
| Validation: 60 | |||||||
| Test: 104 | |||||||
| Sandfort/Summers 2019 (Sandfort et al. [54]) | USA | Gastroenterology | CycleGAN | Non-contrast image generation to augment task | CT | Total: 419 | Dice: |
| Kidney: 0.66 | |||||||
| Liver: 0.89 | |||||||
| Spleen: 0.69 | |||||||
| Chi/Kumar 2018 (Chi et al. [39]) | Australia | Dermatology | CycleGAN | Melanoma generation to augment task | Dermascopy | Total: 1279 | Dice: 0.844 |
| Train: 900 | |||||||
| Test: 379 | |||||||
| Liu/Jambawalikar 2019 (Liu et al. [55]) | USA | Urology | CycleGAN | Translation of prostate MRI to synthetic prostate CT to augment task | CT MR | Total: 526 | Dice: 0.83 ± 0.13 |
| Train: 346 | |||||||
| Validation: 60 | |||||||
| Test: 120 | |||||||
| Jafari/Abolmaesumi 2019 (Jafari et al. [48]) | Canada | Cardiology | Vanilla GAN | Translation of cardiac ultrasounds to segmentation maps | Ultrasound | Dice: | |
| Total: 648 | ED: 0.941 ± 0.033 | ||||||
| Train: 518 | RF: 0.936 ± 0.038 | ||||||
| Test: 130 | ES: 0.93 ± 0.039 | ||||||
| Dong/Xing 2018 (Dong et al. [56]) | USA | Cardiology | Vanilla GAN | Translation of chest X-rays to segmentation maps | X-ray | Total: 247 | Acc: 0.8778 |
| Train: 198 | Prec: 0.9772 | ||||||
| Test: 49 | Sen: 0.8421 | ||||||
| Spec: 0.9557 | |||||||
| Ning/Zhang 2018 (Ning et al. [26]) | China | Gastroenterology | Pancreas-GAN | Translation of pancreatic CT to segmentation maps | CT | Total: 80 | Dice: 88.72 ± 3.23 |
| Train: 60 | |||||||
| Test: 20 | |||||||
| Shin/Michalski 2018 (Shin et al. [57]) | USA | Neurology | pix2pix | Brain tumor generation to augment task | MR | Total: 3416 | Dice: 0.86 ± 0.08 |
| Train: 3416 | |||||||
| Xue/Huang 2018 (Xue et al. [58]) | USA | Neurology | SegAN | Translation into segmentation maps | MR | Total: 274 | Dice: 0.85 |
| Train: 247 | |||||||
| Test: 27 | |||||||
| Xing/Tan 2020 (Xing et al. [27]) | China | Senology | SPCGAN | Semi-pixel-wise translation into segmentation maps | Ultrasound | Total: 640 | Dice: 0.92 ± 0.04 |
| Train: 399 | |||||||
| Validation: 141 | |||||||
| Test: 100 | |||||||
| Huo/Landman 2018 (Huo et al. [29]) | USA | Gastroenterology Neurology | SynSeg-Net | Transfer learning and translation to segmentation map | CT MR | Total: 5136 | Dice: 0.872 ± 0.064 |
| Train: 3262 | |||||||
| Test: 1874 | |||||||
| Enokiya/ Han 2018 (Enokiya et al. [59]) | Japan | Hepatology | Wasserstein GAN | Translation into segmentation maps | CT | Total: 416 | Dice: 0.94 |
| Train: 396 | |||||||
| Test: 20 | |||||||
| Tu/He 2019 (Tu et al. [60]) | China | Ophthalmology | Wasserstein GAN | Translation into segmentation maps | Retinal | Total: 60 | Acc: 0.9571 |
| Train: 30 | Sen: 0.7840 | ||||||
| Test: 30 | Spe: 0.9850 | ||||||
| AUC: 0.9850 | |||||||
| Neff/Urschler 2018 (Neff et al. [61]) | Austria | Pulmonology | Wasserstein GAN | Chest X-ray generation to augment task | X-ray | Total: 247 | Dice: 0.9712 |
| Train: 135 | |||||||
| Validation: 30 | |||||||
| Test: 82 | |||||||
| Shi/Li 2020 (Shi et al. [62]) | China | Pulmonology | AUGAN | Translation into segmentation maps | CT | Total: 4709 | Dice: 0.869 |
| Train: 3326 | |||||||
| Test: 1413 | |||||||
| Decourt/Duong 2020 (Decourt and Duong [63]) | France | Cardiology | DT-GAN | Segmentation of the left ventricle | MRI | Total: 5011 | Dice: 0.88 ± 0.08 |
| Lei/Wang 2020 (Lei et al. [64]) | China | Dermatology | DAGAN | Skin lesion segmentation | Dermascopy | Total:2596 | Acc: 0.935, |
| Train: 2296 | Sen: 0.835 | ||||||
| Test: 300 | Spec: 0.976 | ||||||
| Dice: 0.859 | |||||||
| Shi/Zhou 2020 (Shi et al. [65]) | China | Pulmonology | style based GAN | Generate images to augment data | CT | Total: 1010 | Dice: 85.21% |
| Hamghalam/Lei 2020 (Hamghalam et al. [66]) | China | Neurology | CycleGAN | Generates HTC images to augment segmentation | MRI | Total: 2000 | Dice: + 0.8% |
| + 0.6%, + 0.5% | |||||||
| Zhao/Shen 2018 (Zhao et al. [67]) | USA | Surgery | Deep-supGAN | Segment bony structures from MRI and CT | MRI, CT | Total 16 | Dice: 0.9446 |
| Yu/Zhang 2019 (Yu et al. [68]) | China | Cardiology | SC-GAN | Vessel segmentation | Angiography | Total: 1092 | Dice: 0.824 ± 0.026 |
| Train: 546 | |||||||
| Test: 218 | |||||||
| Val: 328 | |||||||
| Shi/Xu 2020 (Shi et al. [69]) | China | Cardiology | cGAN | Segmentation mask generation | Angiography | Total: 3873 | CRA Acc: 83.53 |
| Train: 3486 | RIGHT Acc: 80.47 | ||||||
| Test: 387 | |||||||
| Kugelman/Collins 2019 (Kugelman et al. [70]) | Austraila | Ophthalmology | GAN | Generate images to augment data | CT | Total: 99 | SCI MAE: 5.82 |
| ILM MAE 0.59 | |||||||
| RPE MAE:0.51 | |||||||
| Hamghalam/Lei 2020 (Hamghalam et al. [71]) | China | Neurology | Enh-Seg-GAN | Generate high contrast images to segment | MRI | Total: 40 | |
| Train: 30 | Dice: 0.89 | ||||||
| Test: 10 | Sen: 0.96 | ||||||
| Dai/Xing 2018 (Dai et al. [72]) | USA | Pulmonology | SCAN | Segmentation mask generation | X-ray | Total: 247 | Dice: 97.3 ± 0.2% |
| Train: 209 | |||||||
| Test: 38 | |||||||
| Xu/Li 2019 (Xu et al. [73]) | Canada | Cardiology | PSCGAN | Generate and segment heart disease | MRI | Total: 280 | RMSE:0.14 |
| Acc: 97.17% |
Acc accuracy, Sen sensitivity, Spe specificity, Prec precision, Dice Dice similarity coefficient, PSNR peak-signal-to-noise ratio, SSIM structural similarity index, MS-SSIM multi-scale SSIM
Evidence Synthesis
Figure 1 presents a flowchart of the study screening and selection process. After removing duplicates and excluding studies based on title and abstract using our study selection criteria, 1206 studies remained for full-text screening. A total of 54 studies fulfilled our eligibility criteria and were included for systematic review and data extraction by our internal reviewers. To measure the consistency between the reviewers, we performed an inter-reviewer assessment study with 20 randomized papers, 10 selected, and 10 rejected from the combined list. To assess the inter-reviewer agreement across the reviewers, we calculated the Fleiss’ Kappa statistic, a well-known and widely used index of the reliability of agreement across multiple reviewers [38]. Across reviewers, a Fleiss’ Kappa of 86.67% was achieved for a significantly strong inter-reviewer agreement. Tables 2–4 show a comprehensive summary of the selected papers for review, stratified by task, and the following subsections detail the benchmarking criteria.
Novelty of GAN Architectures
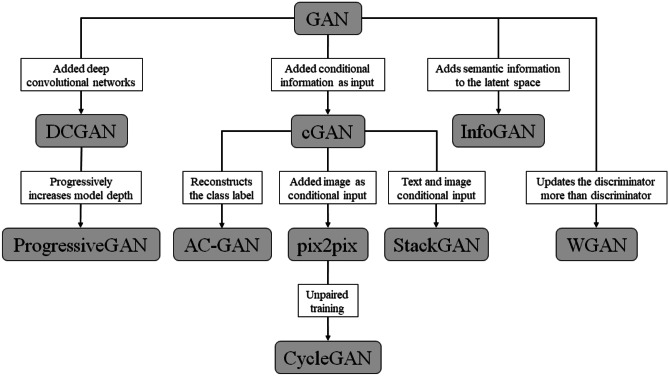
The most common variants of the vanilla GAN architecture used in medical images are (i) cGAN [19], (ii) DCGAN [22], and (iii) pix2pix [24]. Each variant of GANs and their key contributions to the vanilla GAN architecture is summarized in Table 1, and the hierarchical representation with modification to original GAN architecture is shown in Fig. 2. The representative models in medical imaging and their architectures have been discussed in supplemental materials.
Fig. 2.
The hierarchy of various GAN architectures
Recently, many variants of GANs are reportedly applied in medical images, with small and large modifications to the common variants mentioned while maintaining the overall adversarial network architecture. One common modification in these architectures is translating the 2D model to 3D to accommodate 3D radiological scans (e.g., CT, MR) instead of the traditional 2D images. These papers modified the GANs or variants of them to consider the 3D losses and generate 3D volumes of the target images like 3D GAN (35) and 3D cGAN (36). Others used different loss functions, metrics, and architectural modifications to optimize and stabilize the network like Wasserstein GAN (37) and progressive growing GAN (PGGAN) (27) or for specific tasks like confidence GAN (26) for segmentation and fixed-point GAN (30) for generation. Nineteen papers in our review had unique names for their GAN models, but architecture-wise they were close variants of one of the three main variants GANs. From Table 1, we mapped the variant architectures to the representative GAN names by task and report them in Fig. 3b. Overall, most reported architectures share the same general adversarial network architecture and have similar implementation details to other GAN variants with small modifications for their tasks like encoding additional target characteristics for the generated images (38) or being tissue specific (28). However, some architectures introduce highly unique modifications such as fixed-point GAN (30) where the architecture can generate unique and diverse images while preserving “fixed points” such as backgrounds.
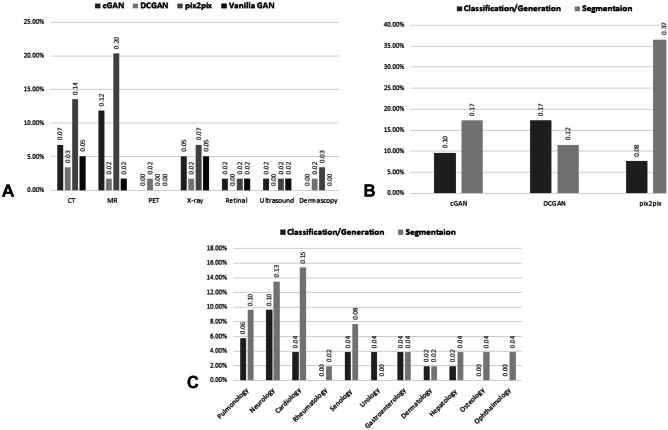
Fig. 3.
Multi-dimensional meta-analysis-distributions of the GAN publications over various factors: a publication counts stratified by imaging modality and GAN architectures. b Publications stratified by targeted task and GAN architectures. c Publications stratified by tasks and clinical domains
Targeted modality
Starting from 2017, GANs have been applied to various medical imaging modalities including screening modalities, like x-ray (mammography and chest), ultrasound (US), retinal fundus images, dermoscopic images, and diagnostic imaging, e.g., magnetic resonance (MR), computed tomography (CT), and positron emission tomography (PET) images. The two most common imaging modalities that GANs were applied to were MR with 21 studies and CT with 17 studies, each primarily focusing on the brain/chest and chest/abdomen regions respectively. Although a significant number of x-ray screening exams are performed every year in radiology departments, it was the third most common modality with 11 studies focusing on the chest/abdomen and mammographic images. Finally, ultrasound, dermoscopic, PET, and retinal fundus images were in the minority with a few studies each. This could be due to limited availability of open-source datasets in these domains and the complexity of processing the images. Sample sizes of each modalities varied based on the availability of the data and dataset used to train the GAN, ranging from over 100,000 chest X-rays (50) and 38,000 MRIs (40), to as little as 50 echocardiograms (41).
Medical Task
The distribution of the tasks within each modality was fairly uniform between GANs for image segmentation and GANs for image augmentation (for increasing training size for classification and segmentation tasks). However, the distribution of tasks within the total set of reviewed papers was heavily skewed with 33 papers (60%) on GAN segmentation (21 papers of direct GAN segmentation and 12 papers augmenting segmentation tasks with GANs), 13 (24%) papers for GAN generation, and 9 (16%) on GAN augmentation of classification. In the 21 papers on GAN augmentation for classification and segmentation, very few papers reported the generated augmentations with Chi et al. [39] and Shen et al. [40] reporting experiments with exactly half (50%) and a little more than half (54%) of the training set being GAN generated images respectively. We discussed the targeted tasks by dividing into the following two subsections: GAN generation/classification and GAN segmentation.
GAN Generation and Augmentation for Classification
A total of 24% papers in Tables 2 and 3 used GANs only to generate artificial medical images without any follow-up targeted. In such cases, the quality of the generated images was measured in both subjective and objective scale. Subjective measures of generated image quality were visual inspections as well as true positive and negative detection rates of several radiologists when presented with real and generated images. Objective measures included comparison of image features of the real and generated lesions such as peak-signal-to-noise ratio (PSNR) and structural similarity index (SSIM). In most cases, GANs successfully generate realistic images with low PSNR and high SSIM. A total of 16% of the papers used GANs to generate more images to augment imbalanced classes for classification to increase the training size and increase the minority class which is usually diseased images. They reported a combination of the final test set classification metrics such as accuracy, sensitivity, specificity, and area-under the receiver-operating-curve (AUC). Across the board, high metrics were reported; Yang et al. [41] reported an GAN augmented classification accuracy, sensitivity, specificity, and AUC of 0.9171, 0.5833, 0.9774, and 0.8812, respectively. When compared to the training results without GAN augmentation, there were improvements in overall metrics.
Table 3.
Benchmarking of the 9 selected manuscripts on GAN classification
| Authors (first/last) | Country | Clinical | GAN | Application | Imaging | Sample | Performance |
|---|---|---|---|---|---|---|---|
| Year | Domain | Architecture | Modality | Sizes | |||
| Yang/Comaniciu 2019 (Yang et al. [41]) | USA | Pulmonology | 3D cGAN | Lung nodule generation to augment task | CT | Total: 1562 | Acc: 0.9171 |
| Train: 1249 | Sen: 0.5833 | ||||||
| Validation: 156 | Spe: 0.9774 | ||||||
| Test: 157 | AUC: 0.8812 | ||||||
| Doman/Mekada 2020 (Doman et al. [42]) | Japan | Hepatology | DCGAN | Liver lesion generation to augment task | CT | Total: 126 | Detection |
| Train: 106 |
Rate: 0.95 False Detection Rate: 0.20 |
||||||
| Test: 20 | |||||||
| Bhattacharya/Mitra 2020 (Bhattacharya et al. [43]) | India | Pulmonology | DCGAN | Lung disease generation to augment task | X-ray | Total: 27,524 | Acc: 0.653 |
| Train: 23,524 | |||||||
| Validation: 2000 | |||||||
| Test: 2000 | |||||||
| Li/Laurenson 2019 (Li et al. [20]) | UK | Senology | DiagNet | Breast mass generation to augment task | X-ray | Total: 107 | Acc: |
| Train: 86 | 0.934 ± 0.019 | ||||||
| Test: 21 | AUC: 0.950 ± 0.02 | ||||||
| Frid Adar/Greenspan 2018 (Frid-Adar et al. [44]) | Israel | Hepatology | Vanilla GAN | Generate liver lesions to augment task | CT | Total: 182 | Sen: 0.857 |
| Train: 63 | Spe: 0.924 | ||||||
| Validation: 63 | |||||||
| Test: 62 | |||||||
| Kim/Ro 2018 (Lee et al. [13]) | South Korea | Senology | Vanilla GAN | Generate breast masses to augment task | X-ray | Total: 960 | AUC: 0.908 |
| Train: 768 | |||||||
| Test: 192 | |||||||
| Han/Hayashi 2019 (Han et al. [23]) | Japan | Neurology | PGGAN | Brain tumor generation to augment task | MR | Total: 12,979 | Acc: 0.9108 |
| Train: 8889 | Sen: 0.8660 | ||||||
| Validation: 1433 | Spec: 0.9760 | ||||||
| Test: 2657 | |||||||
| Kaur/Rani 2020 (Kaur et al. [45]) | India | Neurology | GAN | Generate images to augment data | MRI | Total: 185 | AUC 88.4% |
| Sedigh/Masouleh 2019 (Sedigh et al. [46]) | Iran | Dermatology | GAN | Generation to augment data | Dermascopy | Total: 97 | Acc: + 18% |
Acc accuracy, Sen sensitivity, Spe specificity, Prec precision, Dice Dice similarity coefficient, PSNR peak-signal-to-noise ratio, SSIM structural similarity index, MS-SSIM multi-scale SSIM
Improving Segmentation with GAN Augmentation and Translation
Exactly 60% of the papers in Table 4 used GANs for medical image segmentation task. A total of 21 applied GANs to directly translate the image into a segmentation map, while 12 used GAN generated images to augment training of segmentation models. The papers that used generated images to augment the training of segmentation models reported the final test set segmentation metrics of Dice-similarity coefficient (Dice) and sometimes reported the segmentation pixel accuracy, precision, sensitivity, and specificity. High metrics were reported in several papers, for example, lung nodule segmentation with GAN augmentation. Jin et al. [47] reported a Dice of 0.989 and cardiac ultrasound segmentation with GAN translation. Jafari et al. [48] reported a Dice of 0.941 ± 0.033, 0.936 ± 0.038, and 0.930 ± 0.039 for ED (end-diastolic), RF (random middle frame), and ES (end-systolic) respectively.
Discussion
In this paper, we performed a systematic review of GAN for medical images based on PRISMA guidelines in “Evidence Synthesis”, and in the following subsections, we perform a multi-dimensional meta-analysis of the reported studies and discuss the limitations and reliability of the review.
Meta-analysis by Modality
GANs have been popular on smaller datasets, including those private, institutional datasets with less than few hundred cases only since it can alleviate the limitations imposed of smaller dataset size, making them suitable for training complex deep learning based models. Other than the representative variants of GANs, there are many other variants of GAN such as ScarGAN [12], fixed-point GAN [28], and Pancreas-GAN [26]. We observed that most of the uniquely named architectures modified the representative variants’ GAN architecture. These modifications include the transformation of a 2D cGAN architecture into a 3D cGAN [47], unique stacking, and the addition and transition of DCGAN layers into progressive growing GANs [23], and use of different loss functions like Wasserstein loss to stabilize the training of traditional GANs [59]. Figure 3a shows the distribution of papers stratified into the representative variants of GANs as well as vanilla GANs by image modality. The two most popular variants of GAN were cGANs and pix2pix. The ability to easily modify the vanilla GAN architecture and allowing the generator to learn the coarse features from the whole dataset and finetuning with class labels [19, 74] makes cGANs a popular choice for medical image analysis augmentation task, especially for handling class imbalance issues. Translational GANs like pix2pix architecture are popular with MR and CT images because they have the largest, high resolution, and already segmented datasets useful for segmentation training.
We also noticed that 3D architectures are popular with volumetric images from MR and CT. For example, Yang et al. [41] focused on generating 3D CT lung nodules with specified disease classes, i.e., malignant or benign, in a masked volume in the lung. They used their trained 3D cGAN to triple the imbalanced, malignant class, from the original 233 malignant cases for a total of 696 malignant cases. With their augmentations, their pre-trained 3D ResNet-152 showed a marked improvement in accuracy, specificity, and AUC. In practice, screening tests are performed on large patient populations compared to diagnostic tests. Large screening image datasets such as ultrasound [75] and X-ray [76] are usually widely available; however, such datasets suffer from severe class imbalance because most patients receiving screening tests have a normal exam. Therefore, GANs are mostly used to remedy class imbalance for such modalities to train task specific models. On the other hand, diagnostic tests are usually expensive procedures, performed only when necessary. As such, available datasets of such images are usually small and private. In these datasets, GANs are usually applied to both augment the available data and remedy any class imbalances for model training.
Meta-analysis by Task
The task of GAN image generation is either evaluated though visual inspection and detection of fake images by reviewers and quantitative measures like PSNR, or through the performance of a follow-up task like classification. Similarly, GAN image generation has been applied to augment the performance of a segmentation model, while GAN image translation has been used to directly translate an image to a segmented image. Figure 3b shows the distribution of tasks among selected publications against representative GAN architectures. It is evident that all architectures except for DCGAN are more popular for segmentation than they are for classification. While an adversarial network can be used to generate images, GANs can also be used to transform one image to another image, and this is applied to segmentation tasks where the input image needs to be transformed into its segmentation mask. However, DCGAN and other stabilizing variants are popular for the combined task of classification and image generation as DCGAN can generate high resolution images to give astounding improvements of 0.30 (from 0.65 to 0.95) and 0.70 (from 0.90 to 0.20) in detection rate and false positives rates in the classification of metastatic liver lesions [42]. Additionally, DCGANs were combined with other variants of GANs like cGANs to create deep convolutional conditional GANs [77] and may have been included in other architectures but not explicitly stated in their methods.
Meta-analysis by Clinical Domain
Selected publications cover several different clinical domains, from pulmonology, neurology, hepatology, to dermatology. As shown in Fig. 3c, image segmentation seems to be popular in several domains such as cardiology, senology, and neurology. Overall, the fraction of papers dealing with segmentation is greater than the combined fractions of classification and generation related papers for such fields. This is understandable as domains like cardiology and senology where fast and accurate segmentation of organs and lesions like the heart or breast for quantitative assessment are paramount for diagnosis and treatment planning.
The segmentation accuracy is varied in pulmonology, urology, and gastroenterology images likely due to the large inter- and intra-disease variations within images. The lean towards GAN segmentation may be spurred by general trends in medical research. While classification is important, the accurate segmentation of organs and lesions heavily influence the classification accuracy. In the overall clinical workflow, fast and accurate segmentation precedes classification and is an important step towards personalized medicine [78].
Meta-analysis for Developmental Insights
In order to provide insights on three challenging aspects of GAN implementation, i.e., architecture stability, image handling, and applications of outputs, the authors have reviewed in depth several GAN architectures (vanilla GANs, DCGANs, and cGANs) and evaluated them on the same publicly available RSNA Intracranial Hemorrhage dataset [79].
First, the stability of the GAN architecture depends heavily on variability of hyperparameters used. Unfortunately, in the literature, GAN implementations in medical images do not report the model architecture in detail nor their hyperparameters which limits the reproducibility of experiments. They generally refer to the architectures mentioned in Table 1 with the default hyperparameters from DCGANs [22]. From our implementation, we observed that the more stable an architecture is, the more forgiving the hyperparameters such as learning rates, betas, and decays of the generator and discriminator networks were in the training, e.g., a stable training was achieved over a wider range of learning rates. With an unstable architecture, the hyperparameters must be fine-tuned such for each optimizer so that one network, the weaker network, is preferentially updated more often for a good balance between the generator and discriminator [80]. However, this can be rectified to an extent by stabilizing the architecture with some best practices such as architectural guidelines for DCGANs [22], i.e., using batch normalizations after convolutional transpose layers, to standardize activations from the previous layer and help facilitate the flow of gradients during training. Another way to improve the stability of GAN training is using different activation layers like Scaled Exponential Linear Units (SELU) instead of batch normalization and ReLU/LeakyReLU to combat the vanishing gradient problem. The benefit of SELUs is that it is a self-normalizating activation that is internal, which is faster than the external batch normalization, and makes vanishing gradients and exploding gradients impossible [81].
Second, the input data itself has a significant effect on the architecture, computational load, and stabilization of the GANs. Although the overall shape of the GAN architecture may be the same, going from 2D images to 3D images poses significant challenges in training such as computational load and complexity of generator learning [82]. Preprocessing images like resizing, cropping, and processing a grayscale image to 3 channels for deep learning tasks is common [83], due to the availability of many popular pretrained networks [84] being trained on ImageNet [9] which has three color channels. We have observed that training a GAN using highly preprocessed images (e.g., the top performing solution to the RSNA Intracranial Hemorrhage Detection Challenge in 2019 that preprocessed the grayscale CT image into a 3-channel image that highlighted the brain, subdural, and bone regions, respectively [79]) made the training tremendously unstable as the generator had to learn three sample spaces (at the same time. Overall, we achieved the optimal GAN performance on the RSNA head CT dataset by building a vanilla GANs, with the DCGAN architectural guidelines [22] as well as self-normalizing SELU [81] activations, and applying differential update rules for the generator and discriminator during training [80].
Finally, the applications of the output generated images are a key component of consideration in designing the GAN architectures. Most GAN architectures are trained and tested on 3 channel images with low bit-depth like ImageNet [9] and celebA [10]. However, in medical imaging, there are no color channels and the intensity ranges are significantly larger than the traditional 8-bit images such as CT images’ Hounsfield units (HU) ranging from − 2000 to 2000. While subjectively “good” medical images can be generated, if the goal of the image generation is for clinical use, the inverse transformation of the generated image to the raw HU in CT must be considered carefully as HU values are critical in radiotherapy planning [85]. As such, the GAN architecture must be adjusted to generate one channel images as three-channel generation of a one channel image may not be as simple as an average of the channels or single selection. However, as mentioned above, while highly processed and 3-channel images may lead to unstable training, it might be worth the effort in finetuning hyperparameters as the addition of three-channel images allows for a significant improvement in the desired task [79].
Limitations of the Review
Even though GANs have wide-ranging applications in various fields, the scope of this review is limited to application of GAN for selected medical imaging tasks. We have also limited ourselves to papers published from January 2015 to August 2020, while GANs were formally introduced in 2014. We established a 1-year buffer for the development and testing of meaningful GAN-based models for medical images. To limit the scope of this review, we stopped reviewing papers published after May 2020. We also limited ourselves exclusively to radiological medical images, even though GANs have been used to process cell-phone images of skin or cervix [86]. The scope was also limited by focusing on popular imaging tasks like segmentation, classification, and data generation. Although GANs have been used for other tasks such as super-resolution, image denoising, and image modality translation, at the time our review, these tasks were rare and considered to be under the umbrella of the three main tasks through image translation such as PET, MR, or CT super-resolution Mahapatra et al. [87, 88, 89] and object detection and localization through the differences between the translated images, e.g., brain lesion and pulmonary embolism localization in fixed-point GAN [28].
Reliability of Review
Our review is the first-of-its-kind systematic review focused on medical imaging–related application of GAN. Previously, GANs have been surveyed from the perspective of loss and optimization functions, state-of-the-art identification, super-resolution, image denoising, etc. Publications selected for our review went through two filters. The first filter being a systematic process of searching through keywords among known medical imaging research databases. The second filtering was performed by three reviewers with very high inter-rater reliability, i.e., Fleiss’ kappa of 86.67%. We also requested an independent reviewer to go through our selected publications to resolve any conflict. Therefore, the observations and results collected from our review are highly reliable.
Conclusion
In summary, the use of GANs in medical imaging applications has exponentially risen in the last few years. Over the course of our review, we have focused on the use of different variants of GANs to augment, balance the training of, or improve two main tasks: classification and segmentation. We have summarized their applications by task and description of their usage in Table 5.
Table 5.
Description of medical image task and variants of GANs used in each
| Image Task | Variants of GAN | Usage | |||
|---|---|---|---|---|---|
| Vanilla GAN | cGAN | DCGAN | CycleGAN | ||
| Classification | X | X | X | X | Generation of realistic samples (from noise or input image) to solve the class imbalance or lack of training data problem |
| Segmentation | - | - | - | X | Generation of segmentation masks directly from real images through translation |
Historically, the primary methods of addressing the class imbalance problem or lack of training data within deep learning have been image transformation methods such as flipping, rotation, blurring, and adding noise or increasing the weights of the training images by class. However, while these traditional methods of augmentation increase the training sample space, it does not actively explore the true sample space like GANs. It has been demonstrated in our review that the GAN training and augmentation allow us to explore, interpolate between, and generate realistic but unseen samples. These generated samples still need to undergo strict testing and verification as one of the main concerns for the use of GANs in medical imaging is the realism or veracity of these images in clinical use. Initial studies have shown that GANs can fool radiologists [11], and papers discussed in our review have shown that GAN generated images do improve deep learning models. But the fundamental question of if these generated images that are actually or could be true, unseen samples in the real space remain. As such, more extensive verification of generated GAN images, other than visual verification by an expert, must be inquired for the state-of-the-art GAN methods for medical images such as testing the generalizability of GANs in real, out-of-distribution datasets. However, even with all this in mind, our review has shown that GANs have tremendous potential in addressing many of the problems in medical imaging tasks.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
Not applicable.
Funding
The work is partially supported by the Winship Cancer Institute Pilot$ grant.
Availability of Data and Material
Not applicable.
Code Availability
Not applicable.
Declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Goodfellow I, Pouget-Abadie J, Mirza M, et al.: Generative adversarial nets, In Advances in neural information processing systems, 2014.
- 2.Zhang X, Zhu X, Zhang N, et al.: Seggan: Semantic segmentation with generative adversarial network, In 2018 IEEE Fourth International Conference on Multimedia Big Data (BigMM), 2018, IEEE.
- 3.Li C, Wand M. Precomputed real-time texture synthesis with markovian generative adversarial networks. In European conference on computer vision: Springer; 2016. [Google Scholar]
- 4.Wang X, Gupta A. Generative image modeling using style and structure adversarial networks. In European conference on computer vision: Springer; 2016. [Google Scholar]
- 5.Tzeng E, Hoffman J, Saenko K, et al.: Adversarial discriminative domain adaptation, In Proceedings of the IEEE conference on computer vision and pattern recognition, 2017.
- 6.Wang K, Gou C, Duan Y, et al.: Generative adversarial networks: introduction and outlook, IEEE/CAA Journal of Automatica Sinica, 2017, 4, 588–598.
- 7.Kurach K, Lucic M, Zhai X, et al.: The gan landscape: Losses, architectures, regularization, and normalization, 2018.
- 8.Pan Z, Yu W, Yi X, et al.: Recent progress on generative adversarial networks (GANs): A survey, IEEE Access, 2019, 7, 36322–36333.
- 9.Deng J, Dong W, Socher R, et al.: Imagenet: A large-scale hierarchical image database, In 2009 IEEE conference on computer vision and pattern recognition, 2009, Ieee.
- 10.Liu Z, Luo P, Wang X, et al.: Deep learning face attributes in the wild, In Proceedings of the IEEE international conference on computer vision, 2015.
- 11.Chuquicusma MJ, Hussein S, Burt J, et al.: How to fool radiologists with generative adversarial networks? a visual turing test for lung cancer diagnosis, In 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), 2018, IEEE.
- 12.Lau F, Hendriks T, Lieman-Sifry J, et al.: Scargan: chained generative adversarial networks to simulate pathological tissue on cardiovascular mr scansDeep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, 2018, Springer, pp. 343–350.
- 13.Lee J-H, Tae Kim S, Lee H, et al.: Feature2mass: Visual feature processing in latent space for realistic labeled mass generation, In Proceedings of the European Conference on Computer Vision (ECCV), 2018.
- 14.Yi X, Walia E, and Babyn P: Generative adversarial network in medical imaging: A review, Medical image analysis, 2019, 58, 101552. [DOI] [PubMed]
- 15.Gui J, Sun Z, Wen Y, et al.: A review on generative adversarial networks: Algorithms, theory, and applications, arXiv preprint arXiv:200106937, 2020.
- 16.Wang Z, She Q, and Ward TE: Generative adversarial networks in computer vision: A survey and taxonomy, arXiv preprint arXiv:190601529, 2019.
- 17.Moher D, Liberati A, Tetzlaff J, et al.: Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement, PLoS med, 2009, 6, e1000097. [DOI] [PMC free article] [PubMed]
- 18.Harzing A-W: The publish or perish book, 2010, Tarma Software Research Pty Limited.
- 19.Mirza M, and Osindero S: Conditional generative adversarial nets, arXiv preprint arXiv:14111784, 2014.
- 20.Li H, Chen D, Nailon WH, et al.: Signed Laplacian Deep Learning with Adversarial Augmentation for Improved Mammography Diagnosis, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019a, Springer.
- 21.Nie D, and Shen D: Adversarial Confidence Learning for Medical Image Segmentation and Synthesis, International Journal of Computer Vision, 2020, 1–20. [DOI] [PMC free article] [PubMed]
- 22.Radford A, Metz L, and Chintala S: Unsupervised representation learning with deep convolutional generative adversarial networks, arXiv preprint arXiv:151106434, 2015.
- 23.Han C, Rundo L, Araki R, et al.: Infinite brain MR images: PGGAN-based data augmentation for tumor detectionNeural approaches to dynamics of signal exchanges, 2020, Springer, pp. 291–303.
- 24.Isola P, Zhu J-Y, Zhou T, et al.: Image-to-image translation with conditional adversarial networks, In Proceedings of the IEEE conference on computer vision and pattern recognition, 2017.
- 25.Zhu J-Y, Park T, Isola P, et al.: Unpaired image-to-image translation using cycle-consistent adversarial networks, In Proceedings of the IEEE international conference on computer vision, 2017.
- 26.Ning Y, Han Z, Zhong L, et al.: Automated pancreas segmentation using recurrent adversarial learning, In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2018, IEEE.
- 27.Xing J, Li Z, Wang B, et al.: Lesion Segmentation in Ultrasound Using Semi-pixel-wise Cycle Generative Adversarial Nets, IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2020. [DOI] [PubMed]
- 28.Siddiquee MMR, Zhou Z, Tajbakhsh N, et al.: Learning fixed points in generative adversarial networks: From image-to-image translation to disease detection and localization, In Proceedings of the IEEE International Conference on Computer Vision, 2019. [DOI] [PMC free article] [PubMed]
- 29.Huo Y, Xu Z, Moon H, et al.: Synseg-net: Synthetic segmentation without target modality ground truth, IEEE transactions on medical imaging, 2018, 38, 1016–1025. [DOI] [PMC free article] [PubMed]
- 30.Kwon G, Han C, and Kim D-s: Generation of 3D brain MRI using auto-encoding generative adversarial networks, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019, Springer.
- 31.Ossenberg-Engels J, and Grau V: Conditional Generative Adversarial Networks for the Prediction of Cardiac Contraction from Individual Frames, In International Workshop on Statistical Atlases and Computational Models of the Heart, 2019, Springer.
- 32.Islam J, and Zhang Y: GAN-based synthetic brain PET image generation, Brain Informatics, 2020, 7, 1–12. [DOI] [PMC free article] [PubMed]
- 33.Yu Z, Xiang Q, Meng J, et al.: Retinal image synthesis from multiple-landmarks input with generative adversarial networks, Biomedical engineering online, 2019b, 18, 1–15. [DOI] [PMC free article] [PubMed]
- 34.Zhang T, Fu H, Zhao Y, et al.: SkrGAN: Sketching-rendering unconditional generative adversarial networks for medical image synthesis, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019, Springer.
- 35.Xu Z, Wang X, Shin H-C, et al.: Tunable CT lung nodule synthesis conditioned on background image and semantic features, In International Workshop on Simulation and Synthesis in Medical Imaging, 2019, Springer.
- 36.Zhou Y, He X, Cui S, et al.: High-resolution diabetic retinopathy image synthesis manipulated by grading and lesions, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019, Springer.
- 37.Li H, Paetzold JC, Sekuboyina A, et al.: Diamondgan: unified multi-modal generative adversarial networks for mri sequences synthesis, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019b, Springer.
- 38.Falotico R, and Quatto P: Fleiss’ kappa statistic without paradoxes, Quality & Quantity, 2015, 49, 463–470.
- 39.Chi Y, Bi L, Kim J, et al.: Controlled synthesis of dermoscopic images via a new color labeled generative style transfer network to enhance melanoma segmentation, In 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2018, IEEE. [DOI] [PubMed]
- 40.Shen T, Gou C, Wang F-Y, et al.: Learning from adversarial medical images for X-ray breast mass segmentation, Computer methods and programs in biomedicine, 2019, 180, 105012. [DOI] [PubMed]
- 41.Yang J, Liu S, Grbic S, et al.: Class-aware adversarial lung nodule synthesis in CT images, In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019, IEEE.
- 42.Doman K, Konishi T, and Mekada Y: Lesion Image Synthesis Using DCGANs for Metastatic Liver Cancer DetectionDeep Learning in Medical Image Analysis, 2020, Springer, pp. 95–106. [DOI] [PubMed]
- 43.Bhattacharya D, Banerjee S, Bhattacharya S, et al.: GAN-Based Novel Approach for Data Augmentation with Improved Disease Classification. Advancement of Machine Intelligence in Interactive Medical Image Analysis, 2020, Springer, pp. 229–239.
- 44.Frid-Adar M, Diamant I, Klang E, et al.: GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification, Neurocomputing, 2018, 321, 321–331.
- 45.Kaur S, Aggarwal H, and Rani R: MR Image Synthesis Using Generative Adversarial Networks for Parkinson’s Disease Classification, In Proceedings of International Conference on Artificial Intelligence and Applications, 2020, Springer.
- 46.Sedigh P, Sadeghian R, and Masouleh MT: Generating Synthetic Medical Images by Using GAN to Improve CNN Performance in Skin Cancer Classification, In 2019 7th International Conference on Robotics and Mechatronics (ICRoM), 2019, IEEE.
- 47.Jin D, Xu Z, Tang Y, et al.: CT-realistic lung nodule simulation from 3D conditional generative adversarial networks for robust lung segmentation, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, Springer.
- 48.Jafari MH, Girgis H, Abdi AH, et al.: Semi-supervised learning for cardiac left ventricle segmentation using conditional deep generative models as prior, In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019, IEEE.
- 49.Liao H, Tang Y, Funka-Lea G, et al.: More knowledge is better: Cross-modality volume completion and 3d+ 2d segmentation for intracardiac echocardiography contouring, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, Springer.
- 50.Chaitanya K, Karani N, Baumgartner CF, et al.: Semi-supervised and task-driven data augmentation, In International conference on information processing in medical imaging, 2019, Springer.
- 51.Rezaei M, Harmuth K, Gierke W, et al. A conditional adversarial network for semantic segmentation of brain tumor. In International MICCAI Brainlesion Workshop: Springer; 2017. [Google Scholar]
- 52.Gaj S, Yang M, Nakamura K, et al.: Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks, Magnetic Resonance in Medicine, 2020, 84, 437–449. [DOI] [PubMed]
- 53.Saffari N, Rashwan HA, Herrera B, et al.: On Improving Breast Density Segmentation Using Conditional Generative Adversarial Networks, In CCIA, 2018.
- 54.Sandfort V, Yan K, Pickhardt PJ, et al.: Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks, Scientific reports, 2019, 9, 1–9. [DOI] [PMC free article] [PubMed]
- 55.Liu Y, Khosravan N, Liu Y, et al.: Cross-Modality Knowledge Transfer for Prostate Segmentation from CT ScansDomain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data, 2019, Springer, pp. 63–71.
- 56.Dong N, Kampffmeyer M, Liang X, et al. Unsupervised domain adaptation for automatic estimation of cardiothoracic ratio. In International conference on medical image computing and computer-assisted intervention: Springer; 2018. [Google Scholar]
- 57.Shin H-C, Tenenholtz NA, Rogers JK, et al.: Medical image synthesis for data augmentation and anonymization using generative adversarial networks, In International workshop on simulation and synthesis in medical imaging, 2018, Springer.
- 58.Xue Y, Xu T, Zhang H, et al.: Segan: Adversarial network with multi-scale l 1 loss for medical image segmentation, Neuroinformatics, 2018, 16, 383–392. [DOI] [PubMed]
- 59.Enokiya Y, Iwamoto Y, Chen Y-W, et al.: Automatic Liver Segmentation Using U-Net with Wasserstein GANs, Journal of Image and Graphics, 2018, 6.
- 60.Tu W, Hu W, Liu X, et al.: DRPAN: A novel Adversarial Network Approach for Retinal Vessel Segmentation, In 2019 14th IEEE Conference on Industrial Electronics and Applications (ICIEA), 2019, IEEE.
- 61.Neff T, Payer C, Štern D, et al.: Generative adversarial networks to synthetically augment data for deep learning based image segmentation, In Proceedings of the OAGM workshop, 2018.
- 62.Shi Z, Hu Q, Yue Y, et al.: Automatic Nodule Segmentation Method for CT Images Using Aggregation-U-Net Generative Adversarial Networks, Sensing and Imaging, 2020c, 21, 1–16.
- 63.Decourt C, and Duong L: Semi-supervised generative adversarial networks for the segmentation of the left ventricle in pediatric MRI, Computers in Biology and Medicine, 2020, 123, 103884. [DOI] [PubMed]
- 64.Lei B, Xia Z, Jiang F, et al.: Skin Lesion Segmentation via Generative Adversarial Networks with Dual Discriminators, Medical Image Analysis, 2020, 101716. [DOI] [PubMed]
- 65.Shi H, Lu J, and Zhou Q: A Novel Data Augmentation Method Using Style-Based GAN for Robust Pulmonary Nodule Segmentation, In 2020 Chinese Control And Decision Conference (CCDC), 2020a, IEEE.
- 66.Hamghalam M, Wang T, and Lei B: High tissue contrast image synthesis via multistage attention-GAN: Application to segmenting brain MR scans, Neural Networks, 2020a. [DOI] [PubMed]
- 67.Zhao M, Wang L, Chen J, et al.: Craniomaxillofacial bony structures segmentation from MRI with deep-supervision adversarial learning, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2018, Springer. [DOI] [PMC free article] [PubMed]
- 68.Yu F, Zhao J, Gong Y, et al.: Annotation-free cardiac vessel segmentation via knowledge transfer from retinal images, In International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019a, Springer.
- 69.Shi X, Du T, Chen S, et al.: UENet: A Novel Generative Adversarial Network for Angiography Image Segmentation, In 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2020b, IEEE. [DOI] [PubMed]
- 70.Kugelman J, Alonso-Caneiro D, Read SA, et al.: Constructing synthetic chorio-retinal patches using generative adversarial networks, In 2019 Digital Image Computing: Techniques and Applications (DICTA), 2019, IEEE.
- 71.Hamghalam M, Wang T, Qin J, et al.: Transforming Intensity Distribution of Brain Lesions Via Conditional Gans for Segmentation, In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 2020b, IEEE.
- 72.Dai W, Dong N, Wang Z, et al.: Scan: Structure correcting adversarial network for organ segmentation in chest x-raysDeep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, 2018, Springer, pp. 263–273.
- 73.Xu C, Xu L, Ohorodnyk P, et al.: Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs, Medical Image Analysis, 2020, 101668. [DOI] [PubMed]
- 74.Goodfellow I: NIPS 2016 tutorial: Generative adversarial networks, arXiv preprint arXiv:170100160, 2016.
- 75.Cortes C, Kabongo L, Macia I, et al.: Ultrasound image dataset for image analysis algorithms evaluationInnovation in Medicine and Healthcare 2015, 2016, Springer, pp. 447–457.
- 76.Jaeger S, Candemir S, Antani S, et al.: Two public chest X-ray datasets for computer-aided screening of pulmonary diseases, Quantitative imaging in medicine and surgery, 2014, 4, 475. [DOI] [PMC free article] [PubMed]
- 77.Suárez PL, Sappa AD, and Vintimilla BX: Colorizing infrared images through a triplet conditional dcgan architecture, In International Conference on Image Analysis and Processing, 2017, Springer.
- 78.Cardenas CE, Yang J, Anderson BM, et al.: Advances in Auto-Segmentation, Seminars in Radiation Oncology, 2019, 29, 185–197. [DOI] [PubMed]
- 79.Flanders AE, Prevedello LM, Shih G, et al.: Construction of a Machine Learning Dataset through Collaboration: The RSNA 2019 Brain CT Hemorrhage Challenge, Radiology: Artificial Intelligence, 2020, 2, e190211. [DOI] [PMC free article] [PubMed]
- 80.Heusel M, Ramsauer H, Unterthiner T, et al.: Gans trained by a two time-scale update rule converge to a local nash equilibrium, In Advances in neural information processing systems, 2017.
- 81.Klambauer G, Unterthiner T, Mayr A, et al.: Self-normalizing neural networks, In Advances in neural information processing systems, 2017.
- 82.Wu J, Zhang C, Xue T, et al.: Learning a probabilistic latent space of object shapes via 3d generative-adversarial modeling, In Advances in neural information processing systems, 2016.
- 83.Pitaloka DA, Wulandari A, Basaruddin T, et al.: Enhancing CNN with preprocessing stage in automatic emotion recognition, Procedia computer science, 2017, 116, 523–529.
- 84.Paszke A, Gross S, Massa F, et al.: Pytorch: An imperative style, high-performance deep learning library, In Advances in neural information processing systems, 2019.
- 85.Thomas SJ: Relative electron density calibration of CT scanners for radiotherapy treatment planning, Br J Radiol, 1999, 72, 781–786. [DOI] [PubMed]
- 86.Ganesan P, Xue Z, Singh S, et al.: Performance Evaluation of a Generative Adversarial Network for Deblurring Mobile-phone Cervical Images, In 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2019, IEEE. [DOI] [PubMed]
- 87.Mahapatra D, Bozorgtabar B, and Garnavi R: Image super-resolution using progressive generative adversarial networks for medical image analysis, Comput Med Imaging Graph, 2019, 71, 30–39. [DOI] [PubMed]
- 88.Song TA, Chowdhury SR, Yang F, et al.: PET image super-resolution using generative adversarial networks, Neural Netw, 2020, 125, 83–91. [DOI] [PMC free article] [PubMed]
- 89.You C, Li G, Zhang Y, et al.: CT Super-Resolution GAN Constrained by the Identical, Residual, and Cycle Learning Ensemble (GAN-CIRCLE), IEEE Trans Med Imaging, 2020, 39, 188–203. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.
Not applicable.