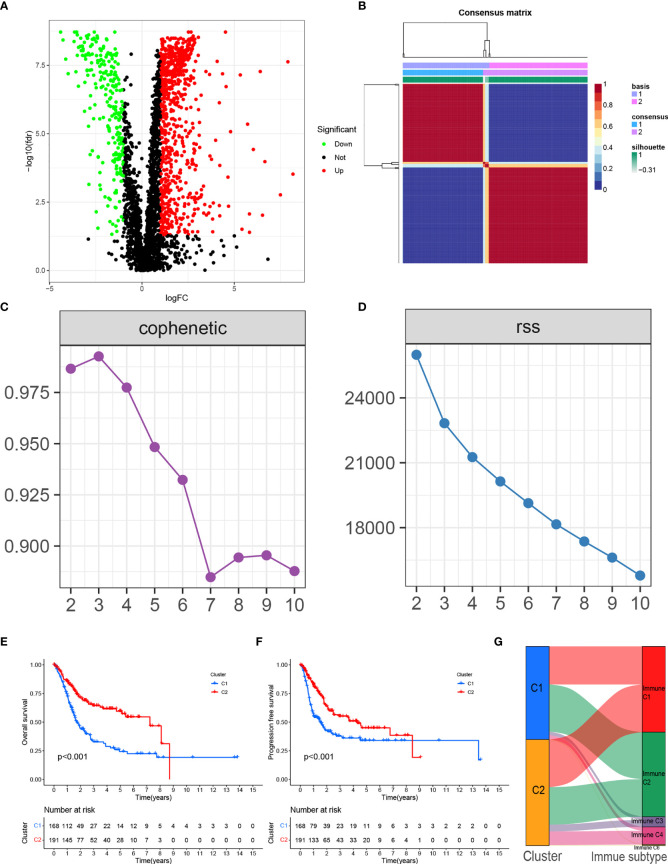
Figure 1.
(A) Volcano plot of differentially expressed genes in BLCA from the TCGA database. (B) Consensus map clustered via the non-negative matrix factorization (NMF) algorithm. (C) The cophenetic correlation coefficient is used to reflect the stability of the cluster obtained from NMF. (D) RSS is used to reflect the clustering performance of the model. (E) Overall survival (OS) showed significant differences between C1 and C2. (F) Progression-free survival (PFS) showed significant differences between C1 and C2. (G) Alluvial plot showing the percentage of C1 and C2 between molecular subtypes.