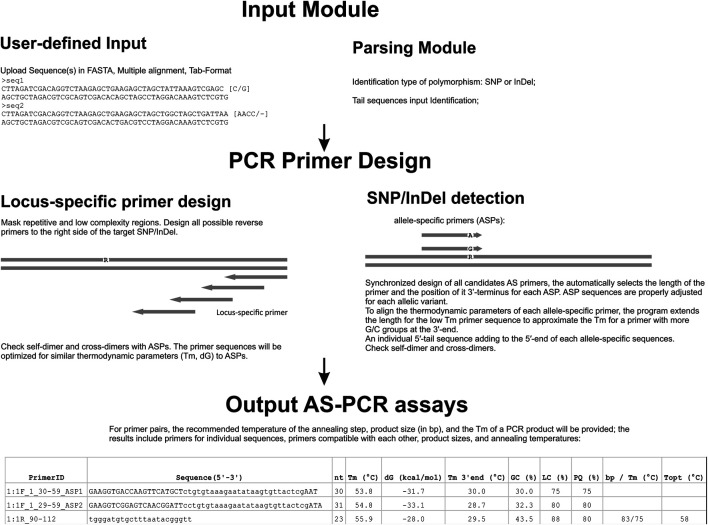
FIGURE 2.
Flowchart of the main steps in the AS-PCR process. Data can be uploaded into FastPCR, which accepts a single sequence or multiple separate DNA sequences in FASTA, tabulated format, multiple alignment, or from clipboard. SNP sequence data are pre-processed by FastPCR prior to execution to collect the SNP position and also to remove non-DNA symbols from the entire sequence. The user sets the various parameters for primer design for the DNA or SNP sequences, such as primer maximum and minimum lengths and T m . Repetitive or low-complexity sequences are excluded from primer design by default (this can be changed by the user). The initial primer design attempts both directions and a reverse primer is designed at the upstream accordingly. An allele-specific primer with the SNP site is designed at the second (penultimate) position of the primer’s 3′ terminus or to any other position in manual or automatic mode. Once the allele-specific primers are designed, an individual 5′-tail sequence is added to the 5′ end of each allele-specific sequence. This combined whole primer sequence is reanalysed for self- and cross-dimerization. Finally, results are returned to the text-editor window.