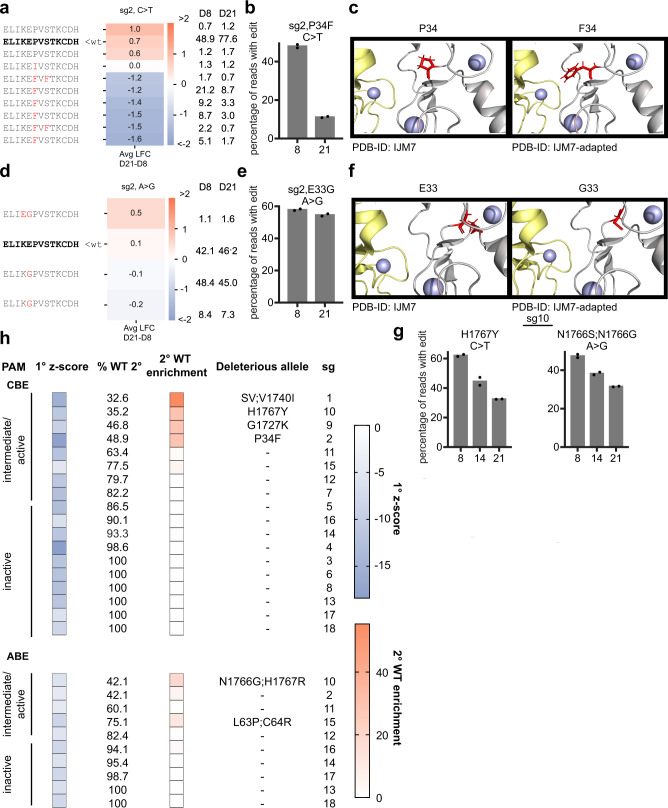
Fig. 6. Validation of BRCA1 hits identified by Cas9-NG.
a Translated sequence around the sgRNA for any allele with at least 1% abundance in any condition. The WT sequence is bolded in black, unchanged amino acids are in grey, and substitutions are highlighted in red. Avg LFC from day 21 to day 8 is indicated on the heatmap and relative percent abundance of each allele is indicated to the right (normalized after filtering for alleles with <1% abundance at both timepoints). b Percentage of all sequencing reads containing the indicated mutation at each timepoint. Dots indicate n = 2 biological replicates. c View of the RING domain (PDB IJM7) of BRCA1 (grey) bound to the RING domain of BARD1 (yellow), with Zn2+ atoms in purple. The left panel shows the canonical amino acid residue in red, the right panel shows the structure with the P34F substitution. d, e Same as (a, b) for sg2 screened with ABE. f Same as (c), but with the E33G substitution. g Same as (b, e). h Summary of validation results. 1° z-score indicates the average z-scored LFC of the sgRNA in the primary screen. % WT 2° indicates the % of reads that were still WT (unedited) on day 8 of the validation experiment. 2° WT enrichment indicates the average change in the abundance of the WT allele from day 8 to day 21 in the validation experiment. PAM bin is indicated on the left. SV indicates “splice variant”. Source data are provided as a Source Data file.