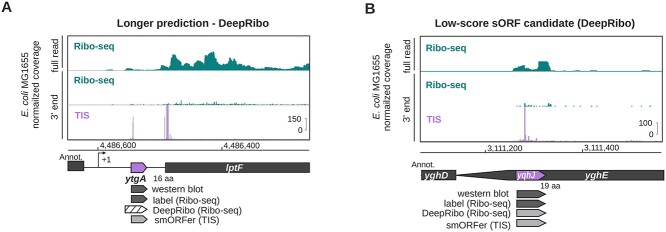
Figure 5.
Detection of novel, western blot-validated E. coli sORFs previously discovered by TIS profiling based. The translation of 31 western blot-validated sORFs previously identified and validated in E. coli [20] was labeled by manual curation of the same Ribo-seq data. Labels were compared with DeepRibo predictions based on the Ribo-seq data at an overlap threshold of 70% based on Ribo-seq data (DeepRibo) or TIS data (smORFer). (A) The sORF ytgA (16 aa), in the 5’UTR of lptF, was labeled as translated and detected in TIS data by smORFer, but the DeepRibo prediction is extended at the 5’ end by three codons. (B) The low-ranked (DeepRibo) sORF yqhJ (19 aa). The ORF was detected by DeepRibo based on Ribo-seq data with a score of -4.078, as well as by TIS profiling by smORFer. Dark gray genes were detected by western blot or labeling. Gray genes were detected by the tools. Hatched arrows were detected, but with a slightly different length or position. White ORFs were not detected. The TIS track was not used for manual curation or DeepRibo predictions and was only used for smORFer. Transcriptional start sites, if available, are indicated with a bent arrow (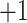 ).
).