Table 1.
Overview of identified ORF detection tools. Most tools make no statement about the taxonomic domain they were developed for. Some, however, utilize eukaryotic data as proof-of-principle (indicated by  ). The first eight tools were benchmarked in this manuscript.
). The first eight tools were benchmarked in this manuscript.
| Name | Input data | Method | Availability | Taxonomy |
|---|---|---|---|---|
| DeepRibo [41] | Ribo-seq | Deep Learning | github | Prokaryotes |
| REPARATION_blast [42] | Ribo-seq | Random Forest | bioconda, github | Prokaryotes |
| SPECtre [37] | Ribo-seq | Spectral Coherence | github | Eukaryotes
|
| Ribo-TISH [36] | Ribo-seq | Negative Binominal Test | bioconda, github | Eukaryotes
|
| IRSOM [21] | RNA-seq | Self-Organizing Map | gitlab, webservice | Eu-, Prokaryotes |
| smORFer [44] | Ribo-seq | Fourier transform | github | Eu-, Prokaryotes |
| PRICE [38] | Ribo-seq | EM-algorithm and statistical testing | github | Eukaryotes
|
| ribotricer [39] | Ribo-seq | 3D to 2D projection for periodicity | bioconda, github | Eukaryotes
|
| RiboTaper [47] | Ribo-/RNA-seq | Multitaper Spectral Analysis | bioconda, galaxy | Eukaryotes
|
| RiboHMM [48] | Ribo-/RNA-seq | Hidden Markov Models | github | Eukaryotes |
| ORFrater [49] | Ribo-seq | Linear Regression | github | Eukaryotes
|
| RibORF [50] | Ribo-seq | Logistic Regression | github | Eukaryotes
|
| Rp-Bp [51] | Ribo-seq | Markov Chain–Monte Carlo | github | Eukaryotes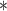
|
