Table 3.
A comprehensive list of the reviewed methods/tools for prediction of prokaryotic and eukaryotic promotersa
| Method type | Toolb | Year | Webserverc | Features/Motifs | Scoring function /Algorithm | Evaluation strategy | Species and promoter type | Sequence length (bp) |
|---|---|---|---|---|---|---|---|---|
| Deep learning–based | CNNProm [30] | 2017 | Yes | – | CNN | 70% train, 20% test, 10% validation |
H. sapiens (TATA-containing and TATA-less), M. musculus (TATA-containing and TATA-less), A. thaliana (TATA-containing and TATA-less), E. coli ( ) and B. subtilis ) and B. subtilis
|
81, 251 |
| Traditional machine learning–based | Rani et al.-I [132] | 2007 | No | DNC | ANN | 5-fold CV and independent test |
E. coli ( ), and D. melanogaster ), and D. melanogaster
|
80, 241 |
| Rani et al.-II [133] | 2009 | No | n-gram (n=2,3,4,5) | ANN | 5-fold CV and independent test |
E. coli ( ) and D. melanogaster ) and D. melanogaster
|
80, 300 | |
| iProEP [31] | 2019 | Yes | PseKNC and PCSF | SVM | 5, 10-fold CV |
D. melanogaster, H. sapiens, C. elegans, E. coli ( ) and B. subtilis ( ) and B. subtilis ( ) ) |
81, 300 | |
| Scoring function–based | IPMD [134] | 2010 | No | PCSF and ID | Modified MD | 10-fold CV and independent test |
D. melanogaster, H. sapiens, C. elegans, E. coli (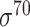 ) and B. subtilis ( ) and B. subtilis (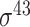 ) ) |
81, 300 |
aAbbreviations: CNN—convolutional neural network; DNC—dinucleotide composition; ANN—artificial neural network; CV—cross-validation; PseKNC—pseudo–K-tuple nucleotide composition; PCSF—position-correlation scoring function; SVM—support vector machine; ID—increment of diversity; MD—Mahalanobis Discriminant.
bThe URL addresses for the predictors with functioning webserver: CNNProm—http://www.softberry.com/berry.phtml?topic=cnnprom&group=programs&subgroup=deeplearn; iProEP—http://lin-group.cn/server/iProEP/pages/download.php.
cYes—The approach is accompanied with a webserver/tool and it is still working; Decommissioned—The webserver/tool is no longer available; No—The approach has no webserver or tool.
