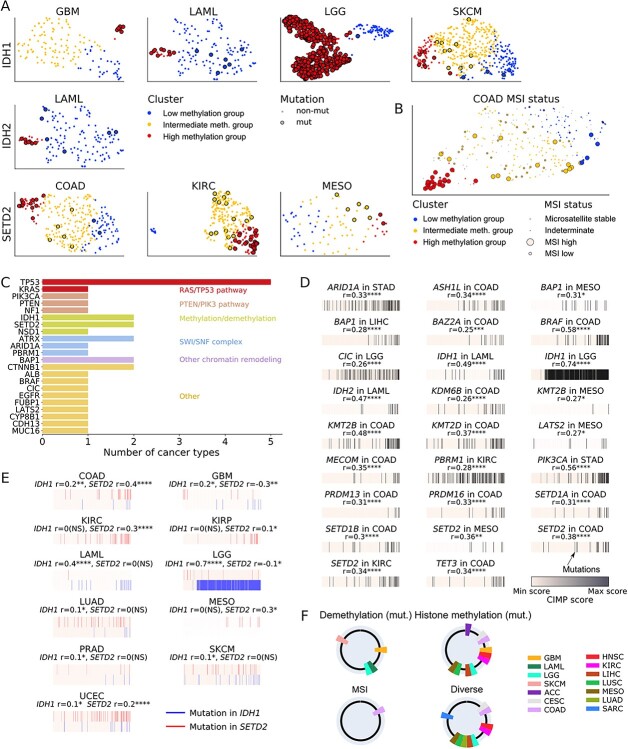
Figure 3.
Discovery of possible drivers of CIMP. (A) UMAP representation of patients with mutations in IDH1/2 or SETD2 genes. (B) MSI status of COAD. MSI annotations are taken from the TCGA consortium calling, using Cortes-Ciriano [55] Supplementary Tables, indeterminate status is colored in gray. (C) Genes selected in the Random Forest analysis as potential drivers for either low, intermediate or high-methylation groups and their associated pathways. We trained Random Forests for 10 CIMP-presenting cancer types. The number of cancer types for which the gene was selected is indicated. (D) Mutations significantly correlated with CIMP score and (E) mutations in IDH1 and SETD2 significantly correlated with CIMP score. Patients are ranked for each cancer type according to their CIMP score. Patients presenting a mutation in the gene of interest are indicated by a black bar, in IDH1 by a blue bar and in SETD2 by a red bar. Point biserial correlation coefficient r is indicated. For (D), only mutations with a correlation coefficient (r) over 0.25 are shown; the full mutation panel is depicted in Supplemental Figure 9. Significance is reported for FDR Benjamini–Hochberg-corrected q-value for (D) and (E). NS: q > 0.1; *: 0.01 ≤ q < 0.1; **: 0.001 ≤ q < 0.01; ***: 0.0001 ≤ q < 0.001; ****: q < 0.0001. (F) Potential etiologies of CIMP in 19 CIMP-positive cancer types. Cancer types are represented as a portion of the circle with an associated color. The four circles represent the four candidate etiologies for CIMP in these cancer types: mutations in the DNA demethylation associated genes, mutations in the histone methylation associated genes, MSI and diverse or unelucidated mechanisms.