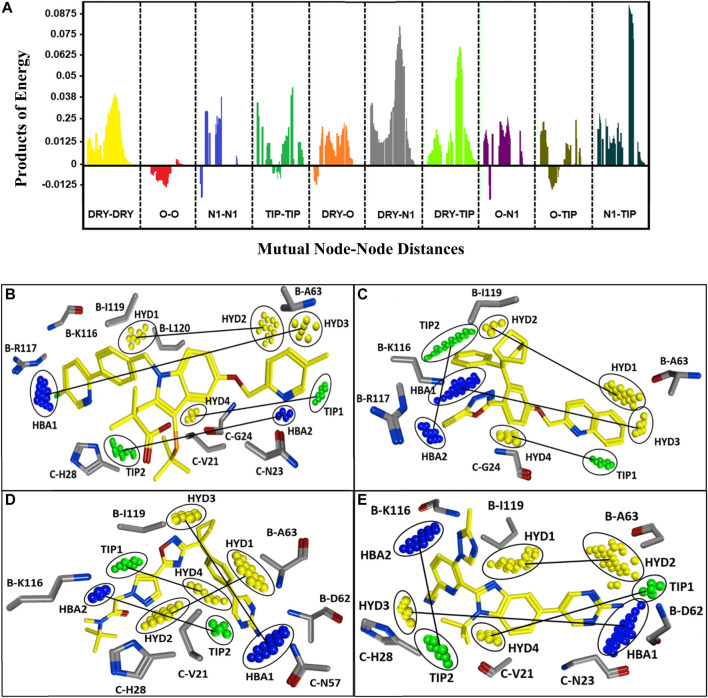
FIGURE 5.
(A) Correlogram of PLS coefficients representing the pair of probes contributing positively (peaks above 0) or negatively (peaks below 0) towards the inhibitory potencies of training set compounds. The positive contribution towards pIC50 of FLAP inhibitors has been depicted by DRY-DRY (two hydrophobic), DRY-N1 (one hydrophobic and one hydrogen bond acceptor), DRY-TIP (one hydrophobic and one steric), and N1-TIP (one hydrogen bond acceptor and one steric) variables. The variables are present in all highly active FLAP compounds and are located at mutual distances of 16.00–16.40 Å, 16.40–16.80 Å, 18.00–18.40 Å, and 17.20–17.60 Å, respectively. (B) The identified hotspots on most active indole-based FLAP inhibitor (compound 1) of training set with projection of actual FLAP structure. Hydrophobic features are depicted in yellow, hydrogen bond acceptors are in blue, while steric hotspots are depicted in green color. The two hydrophobic hotspots (HYD1 and HYD2) are located between two aromatic moieties, one hydrophobic (HYD3) and one hydrogen bond acceptor feature (HBA1) are present between aromatic rings and terminal negative ionizable substitution, one hydrophobic (HYD4) and steric feature (TIP1) can be spotted between aromatic ring and indole scaffold, while one hydrogen bond acceptor (HBA2) and one steric (TIP2) hotspot are present between dimethylbutanoic acid and pyridine ring. (C) The most active compound (compound 10) of class II with mapping of complemented amino acids on the recognized contours. (D) The most active of class III (compound 7), which is also the most active compound from oxadiazole-based FLAP antagonists (classes III, IV, and V) and mapped hotspots along with projection of complementary amino acids of FLAP binding cavity. Due to high structural similarity, the features were also observed at the same positions in all active compounds of classes IV and V. (E) The compound (70) from class VI with identified hotspots and corresponding amino acids.