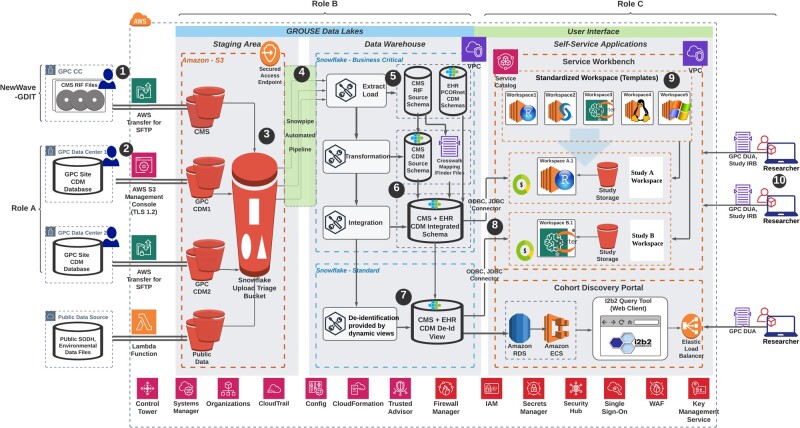
Figure 4.
Data that flow from multiple sources, including (1) NewWave-GDIT physical media and (2) other Greater Plains Collaborative sites will be load into secured S3 bucket via Secure File Transfer Protocol or using AWS S3 management console (TLS 1.2). (3) Raw files are externally staged in S3 buckets and then loaded into Snowflake data warehouse via (4) the Snowpipe automated pipeline (a Snowflake functionality). (5) Data in source Center for Medicare and Medicaid Services (CMS) research identifiable file or site Common Data Model (CDM) schema are first extracted as they are in 1 database. (6) CMS data will then be transformed into PCORnet CDM and integrated with electronic health record data using the finder file provided by CMS. (7) The integrated CDM will be deidentified using the built-in dynamic view functionality provided by Snowflake. (8) Both the limited and deidentified view can be accessed via ODBC or JDBC connector with researchers’ service workbench workspaces. (9) Service workbench provides templated and reusable workspaces (AWS EC2 instances) with various computing power, operating systems and prepackaged software that can satisfy most of the research needs. (10) Approved researchers can deploy the self-serviced applications to perform either advanced analysis using the service workbench or simply discover study cohort using an integrated Informatics for Integrating Biology & the Bedside query tool. Various underlying Amazon Web Services are marked at each step described above as well as at the bottom of the figure.