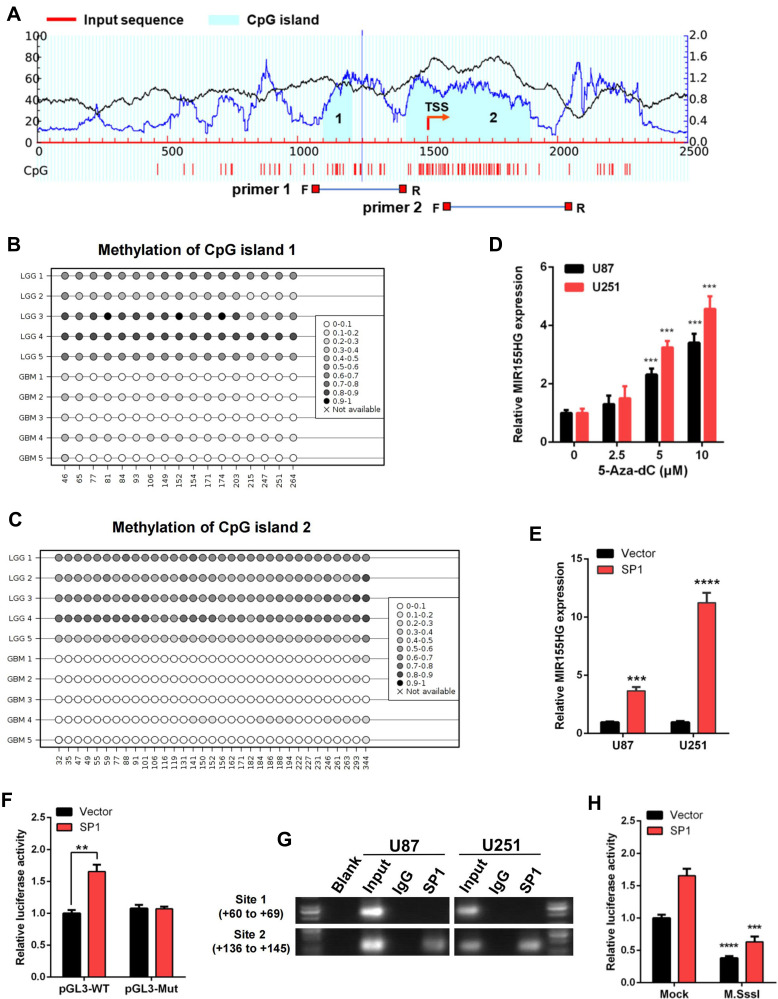
Figure 4.
DNA methylation and SP1 were involved in the activation of MIR155HG. (A) Schematic representation of the CpG islands and bisulfite sequencing primers. (B and C) Bisulfite amplicon sequencing (BSAS) was performed to examine methylation status of CpG island 1 and 2 at the promoter region of MIR155HG in LGG and GBM. (D) MIR155HG expression in U87 and U251 cell lines were upregulated in a dose-dependent manner after 5-Aza-dC treatment. (E) Overexpression of SP1 activated the expression of MIR155HG in U87 and U251 cell lines. (F) The luciferase reporter plasmids carrying MIR155HG promoter pGL3-WT or pGL3-Mut with the SP1 plasmid or vector were co-transfected into HEK293T cells. Relative luciferase activity in HEK293T cells were determined. (G) ChIP assay was performed to confirm the putative binding regions for SP1 in MIR155HG promoter region. (H) Luciferase reporter assay revealed that M.SssI-induced methylation could abolish the MIR155HG promoter activity activated by SP1. **p < 0.01; ***p < 0.001; ****p < 0.0001.