Figure 5. SDSI+AmpSeq is used to identify sample swaps and contamination.
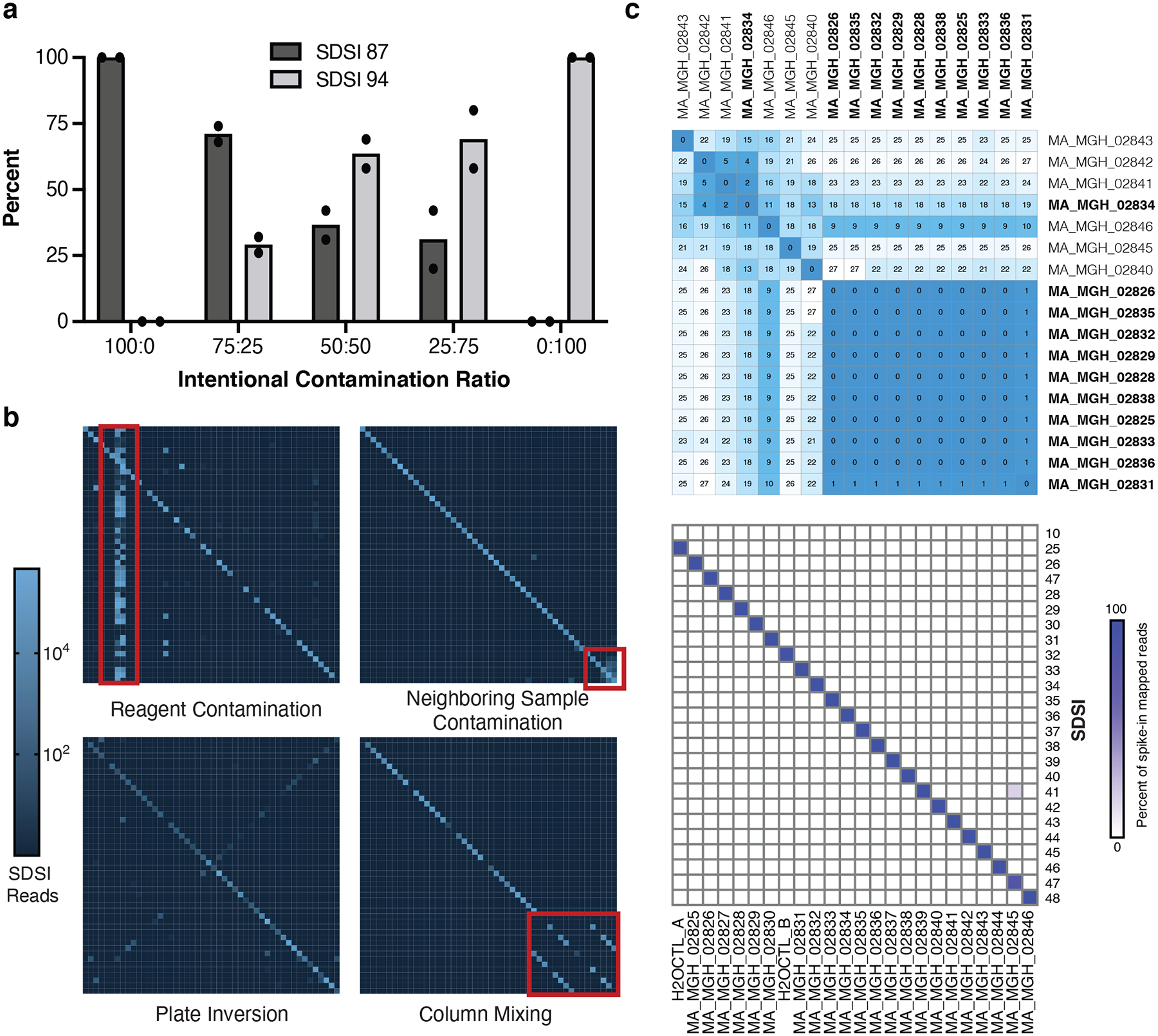
a, Intentional SDSI contamination experiment (run in duplicate) assessing if different ratios of contamination between SDSI 87 and SDSI 94 (SDSI 87:SDSI 94) were detectable with the SDSI+AmpSeq method. b, Examples of experimental errors that were caught using the SDSI+AmpSeq method. c, Top: Distance matrix showing pairwise differences between the 17 complete genomes assembled from this sample set. Putative cluster samples are bolded. Bottom: Spike-in counts for each of the 24 samples and water controls in this sequencing batch.
