FIGURE 1.
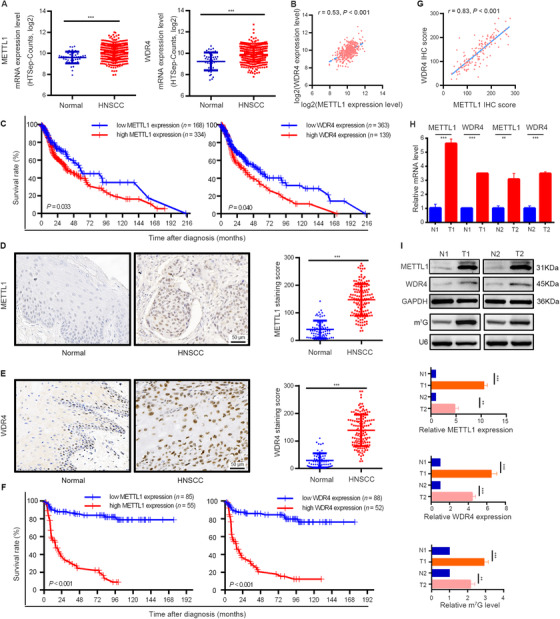
METTL1 and WDR4 are upregulated and associated with poor prognosis of HNSCC patients. (A) TCGA data showed that METTL1 (left) and WDR4 (right) were significantly upregulated in HNSCC tissues (n = 502) as compared with normal tissues (n = 44). (B) The correlation between METTL1 and WDR4 levels in HNSCC tissues using TCGA data. (C) Kaplan‐Meier curves of overall survival based on METTL1 (left) and WDR4 (right) levels in HNSCC tissues from TCGA data. (D) Representative images of IHC staining for METTL1 (left) and quantification (right) between 140 HNSCC tissue samples and 69 normal tissue samples. (E) Representative images of IHC staining for WDR4 (left) and quantification (right) between 140 HNSCC tissue samples and 69 normal tissue samples. (F) Kaplan‐Meier curves of overall survival based on METTL1 (left) and WDR4 (right) levels in HNSCC patients from our cohort. (G) The correlation between METTL1 and WDR4 levels in HNSCC patients from our cohort. (H,I) The mRNA levels and protein levels of METTL1 and WDR4 and m7G tRNA modification were upregulated in human HNSCC specimens examined by qRT‐PCR (H), Western blotting and Northwestern blotting (I). T1/T2 and N1/N2 represented tumor tissues and surrounding normal mucosal tissues from two HNSCC patients, respectively. Data are presented as the mean ± SD and analyzed by Student's t‐test, log rank test, or Pearson's correlation test. **, P < 0.01, ***, P < 0.001. Abbreviations: METTL1: Methyltransferase‐like 1; WDR4: WD repeat domain 4; HNSCC: head and neck squamous cell carcinoma; TCGA: the Cancer Genome Atlas; IHC: immunohistochemistry; m7G: N7‐methylguanosine; qRT‐PCR: quantitative real‐time PCR
