FIGURE 6.
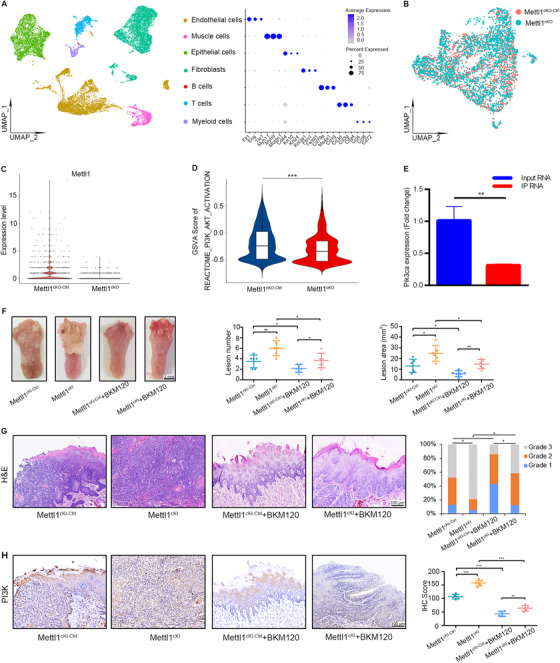
A transgenic mouse model verifies that Mettl1 regulates PI3K/Akt/mTOR activity in HNSCC. (A) UMAP of scRNA‐seq cells recovered from both Mettl1cKO‐Ctrl and Mettl1cKO mice labeled by cell type (left). Dot plot of selected subset‐associated genes across cell types from scRNA‐seq data (right). (B) UMAP of reclustered epithelial cells from Mettl1cKO‐Ctrl and Mettl1cKO mice. (C) Violin plots of the expression of the Mettl1 gene in epithelial cells from Mettl1cKO‐Ctrl and Mettl1cKO mice. (D) Violin plots of the GSVA results for epithelial cells revealed that PI3K/Akt activation was significantly decreased in Mettl1cKO HNSCC samples. (E) qRT‐PCR of Pik3ca expression (K14CreER;RiboTag;Mettl1fl/fl /K14CreER;RiboTag;Mettl1wt/wt ) in input RNA was normalized to that of Gapdh and the ratio of IP RNA. (F) Representative images (left) of tongue lesions from Mettl1cKI‐Ctrl and Mettl1cKI mice by oral gavage of BKM120 (Mettl1cKI‐Ctrl + BKM120 and Mettl1cKI + BKM120) compared with Mettl1cKI‐Ctrl and Mettl1cKI mice by oral gavage of buffer agent (Mettl1cKI‐Ctrl and Mettl1cKI). Quantification of the number of lesions (middle) and the lesion areas (right) visible in the tongue (n = 6). (G) Representative H&E staining of lesions (left) from Mettl1cKI‐Ctrl, Mettl1cKI, Mettl1cKI‐Ctrl + BKM120 and Mettl1cKI + BKM120 mice. Quantification of histological grade of lesions in different groups (right). (H) Representative IHC staining (left) and quantification (right) of PI3K in Mettl1cKI‐Ctrl, Mettl1cKI, Mettl1cKI‐Ctrl + BKM120 and Mettl1cKI + BKM120 mice. Data are presented as the mean ± SD and analyzed by Student's t‐test or Chi‐squared Test. *, P < 0.05, **, P < 0.01, ***, P < 0.001. Abbreviations: PI3K/Akt/mTOR: phosphatidylinositol‐3‐kinase/protein kinase B/mammalian target of rapamycin; Pik3ca: phosphatidylinositol‐4,5‐bisphosphate 3‐kinase, catalytic subunit alpha; Mettl1: Methyltransferase‐like 1; IHC: immunohistochemistry; IP: immunoprecipitation; UMAP: Uniform manifold approximation and projection; GSVA: gene set variation analysis; Mettl1cKO‐Ctrl: K14CreER;Mettl1wt/wt mice; Mettl1cKO: K14CreER;Mettl1fl/fl mice; Mettl1cKI‐Ctrl: K14Cre;Mettl1wt/wt mice; Mettl1cKI: K14Cre;Mettl1KI/KI mice; SD: standard deviation
