FIGURE 7.
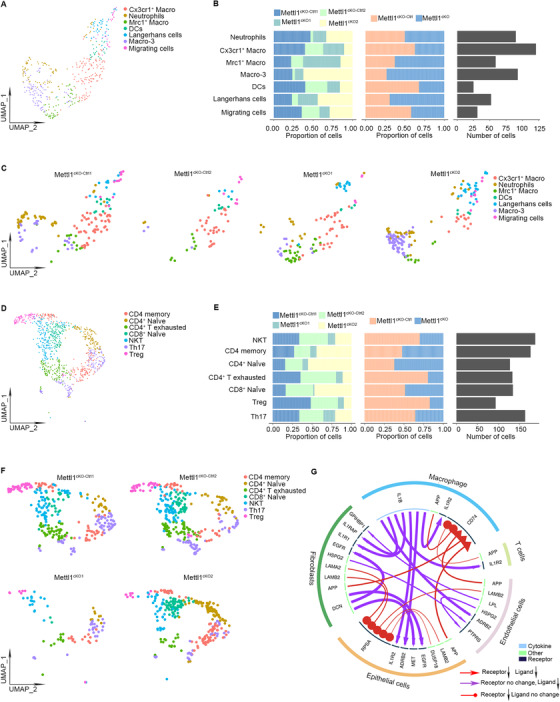
scRNA‐seq analysis of tumor cell‐microenvironment crosstalk in HNSCC tumors after Mettl1 knockout. (A) UMAP of myeloid subsets, including neutrophils, Cx3cr1+ macrophages, Mrc1+ macrophages, Macro‐3, dendritic cells, Langerhans cells, and migrating cells, in Mettl1cKO‐Ctrl and Mettl1cKO mice. (B) Bar plots of the proportions of cell types and total cell number in Mettl1cKO‐Ctrl and Mettl1cKO mice. (C) UMAP of myeloid subsets of each sample. (D) UMAP of TIL subsets, including memory CD4 T cells, naïve CD4+ T cells, exhausted CD4+ T cells, naïve CD8+ T cells, NKT cells, Th17 cells, and Tregs, in Mettl1cKO‐Ctrl and Mettl1cKO mice. (E) Bar plots of the proportions of cell types and total cell number in Mettl1cKO‐Ctrl and Mettl1cKO mice. (F) UMAP of TIL subsets of each sample. (G) Circos plot showing significant crosstalk alterations between stromal cells and epithelial cell subclusters (Mettl1cKO vs. Mettl1cKO‐Ctrl). Abbreviations: Mettl1: Methyltransferase‐like 1; KO: knockout; TIL: tumor‐infiltrating lymphocytes; Mettl1cKO‐Ctrl: K14CreER;Mettl1wt/wt mice; Mettl1cKO: K14CreER;Mettl1fl/fl mice; Macro‐3: macrophage‐3; DCs: dendritic cells; UMAP: Uniform manifold approximation and projection
