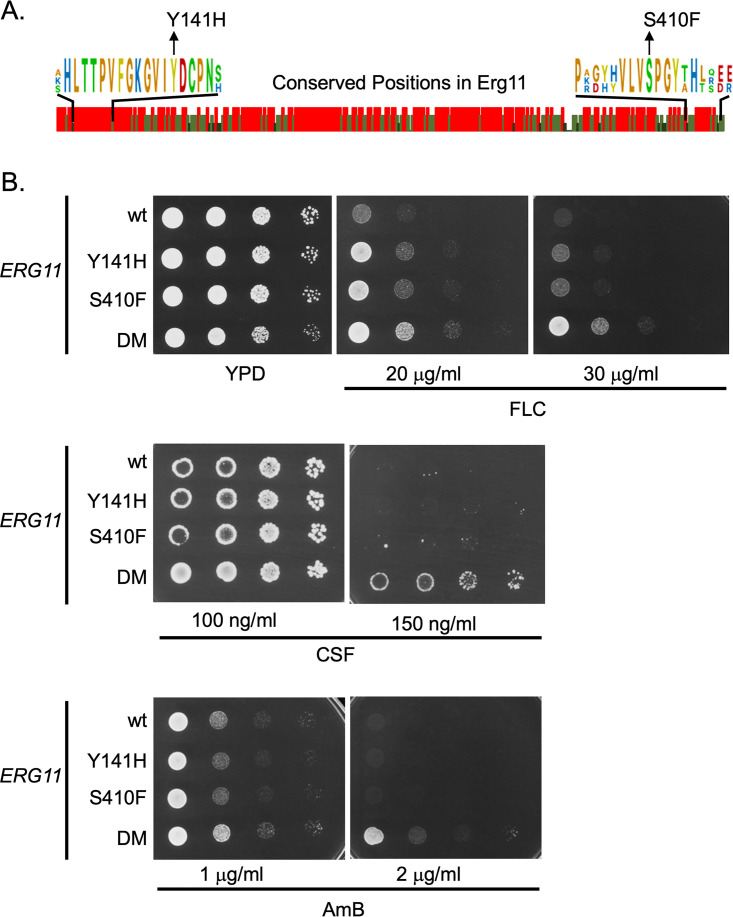
FIG 1.
Construction of mutant ERG11 alleles in Candida glabrata. A. Amino acid sequence comparison between Erg11 proteins from Saccharomyces cerevisiae, Candida glabrata and Candida albicans. Graphic indication of amino acid identity between these three yeast enzymes is shown with a red bar indicating 100%, while light and dark green denotes 66% or no conservation. Expanded regions with conserved residues are shown to illustrate the positions that were mutated in C. glabrata to match changes associated with fluconazole resistance originally found in C. albicans. B. Isogenic C. glabrata strains containing the indicated ERG11 alleles were grown to mid-log phase and plated as serial dilutions on rich medium (YPD) or the same medium containing the indicated concentrations of antifungal drugs. The strain containing both the Y141H- and S410F-encoding alleles was designated double mutant (DM). Plates were allowed to develop at 30°C and then photographed. Antifungal drugs are abbreviated as fluconazole (FLC), caspofungin (CSF) and amphotericin B (AmB).