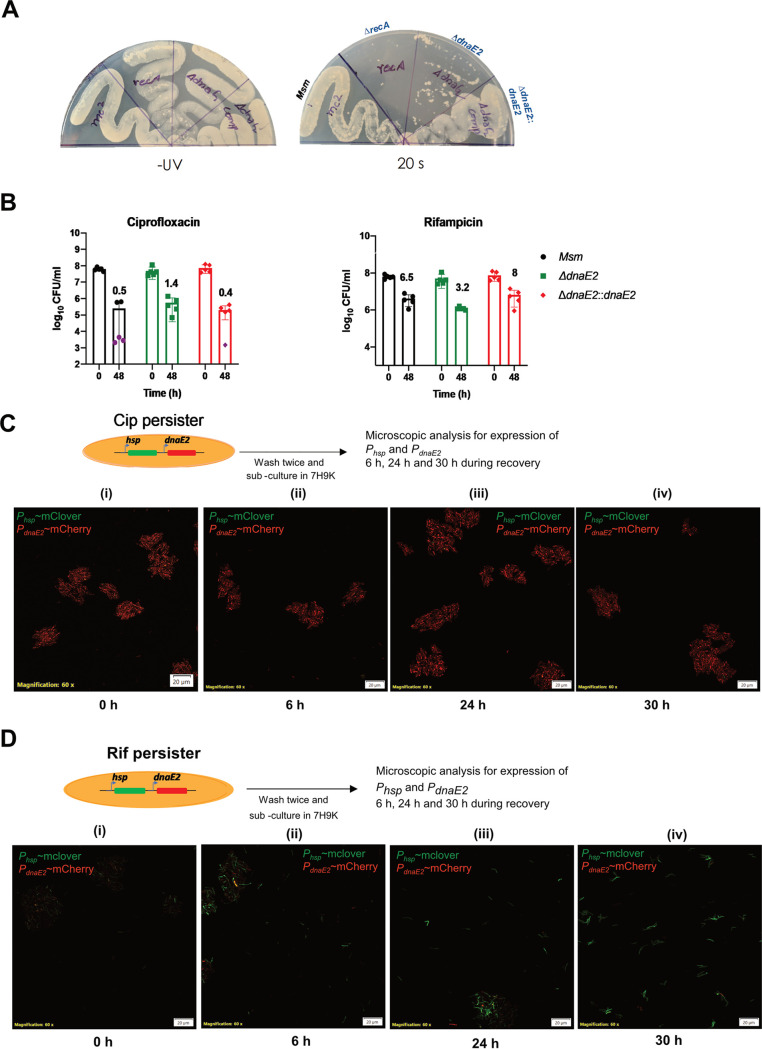
FIG 3.
Characterization of the ΔdnaE2 strain and time course analysis of dnaE2 expression. (A) UV sensitivity analysis. Cultures of M. smegmatis wild-type (Msm), recA::kan, ΔdnaE2, and dnaE2::dnaE2 strains were streaked onto two 7H10 plates, one of which was exposed to UV for 20 s, while the other was left untreated. The plates were incubated at 37°C, and digital images of the plates were captured. (B) Evaluation of bacterial survivors upon antibiotic treatment. Five replicate cultures of M. smegmatis wild-type, ΔdnaE2, and ΔdnaE2::dnaE2 strains were treated with Cip or Rif for 48 h. Scatterplots of surviving bacteria from serial dilution and plating were plotted using GraphPad Prism software, with means ± SD. The average percent survival was determined and is represented above the corresponding bar for each antibiotic. (C and D) Time course expression analysis of dnaE2 was performed by recovering the M. smegmatis dual reporter from either Cip or Rif persister cells. The images were acquired at 0, 6, 24, and 30 h in the recovery stage. The images are representative of results from two independent replicates.