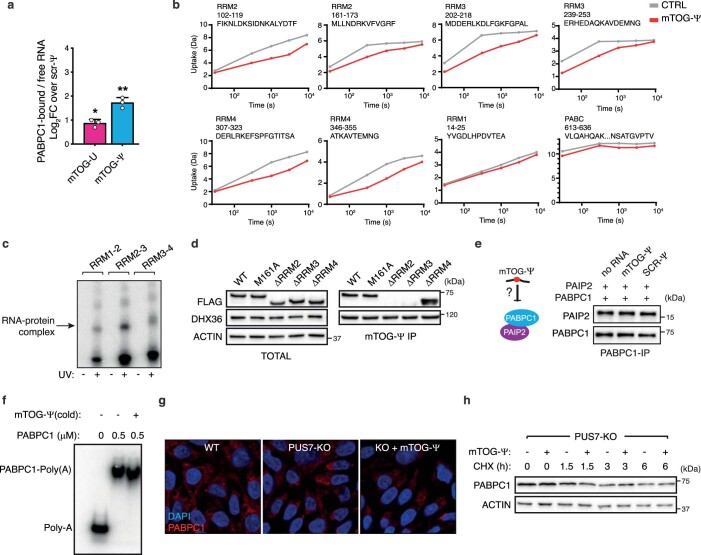
Extended Data Fig. 1. mTOG-Ψ bind to RRM2 and RRM3 within PABPC1.
a, Graph shows fraction of PABPC1 bound to RNA normalized to unbound RNA as the log2 FC to SCR-Ψ oligo in EMSA experiments (n=3 independent experiments, mean ± SD). *p=0.0354; ***p=0.0002 (one-way ANOVA with multiple comparisons). b, Uptake plots show number of deuterium incorporated in selected peptides from indicated PABPC1 domains in HDX-MS time-course experiment in CTR or mTOG-Ψ conditions. Annotations indicate RRM, residue number and peptide sequence. Data points are mean of n=2 biological experiments. c, Binding assay shows significant and selective interaction between mTOG-Ψ and RRM2-3 upon UV-crosslinking. d, Immunoprecipitation experiments using biotinylated mTOG-Ψ oligos in cells transduced with FLAG-tagged full-length or different PABPC1 mutants. DHX36 is shown as a control mTOG-Ψ-bound protein2. e, mTOG-Ψ does not affect PABPC1-PAIP2 interaction. PAIP2 co-precipitated with PABPC1 is shown in the presence or absence of SCR and mTOG-Ψ oligos. f, mTOG-Ψ does not alter PABPC1 binding to poly(A). Representative EMSA shows radioactive-labelled poly(A) RNA incubated with recombinant PABPC1 in presence or absence of cold mTOG-Ψ. g, Immunofluorescence shows no changes in PABPC1 localization in WT and PUS7-KO hESC ± mTOG-Ψ. h, Protein stability analysis show no differences in PABPC1 levels ± mTOG-Ψ in hESCs following treatment with cycloheximide (CHX). Data in c-h represent n=3 independent experiments, except g, showing one representative experiment.