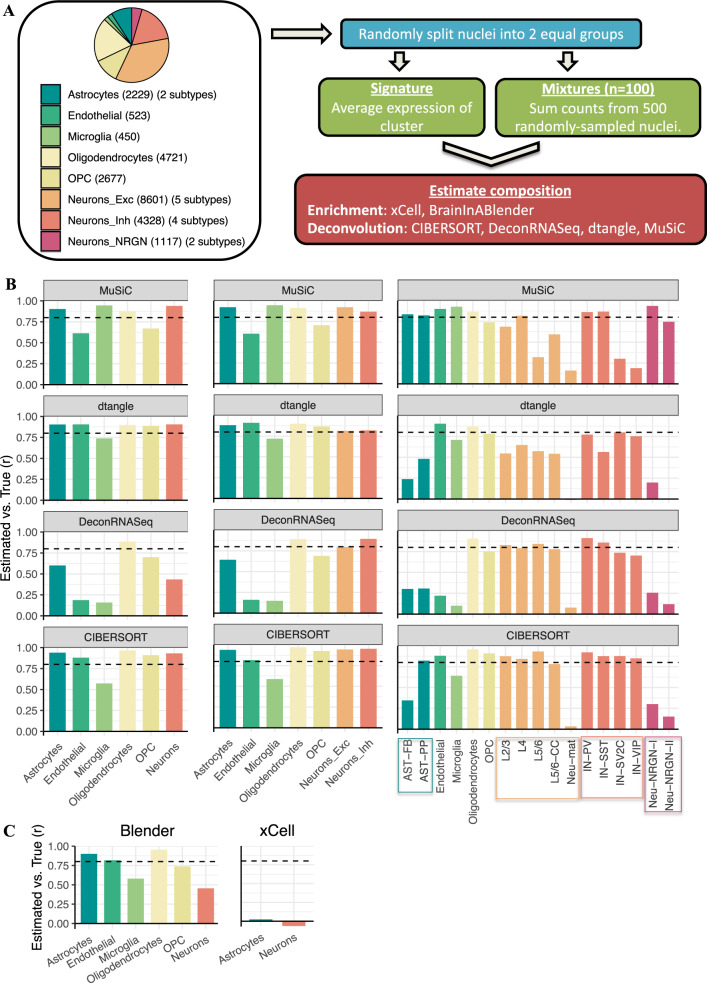
Fig. 1. Deconvolution accuracy across methods.
A Simulation design. Single-nucleus RNA sequencing data was acquired from Velmeshev et al. and used to create 100 in silico mixtures with known proportions (see the “Methods” section). Left: Piechart displaying the composition of the dataset, where n is the number of cells per cell-type. The number of sub-types is listed in between brackets. Right: analysis outline. OPC oligodendrocyte precursor cells. Neurons_Exc, Neurons_Inh, and Neurons_NRGN: excitatory, inhibitory, and NRGN+ neurons, respectively. DRS DeconRNASeq, CIB CIBERSORT, Blender BrainInABlender. B Barplots of Pearson correlation coefficients (r) between true and estimated cell-type proportions in 100 in silico mixtures. Left: cells are grouped by major cell-types; middle: excitatory and inhibitory neuron subtypes are included in the signature; right: all cell-subtype labels are used in the signature. C Barplots of Pearson correlation coefficients (r) between true proportion and cell-type enrichment scores. Dotted horizontal lines: r = 0.8.