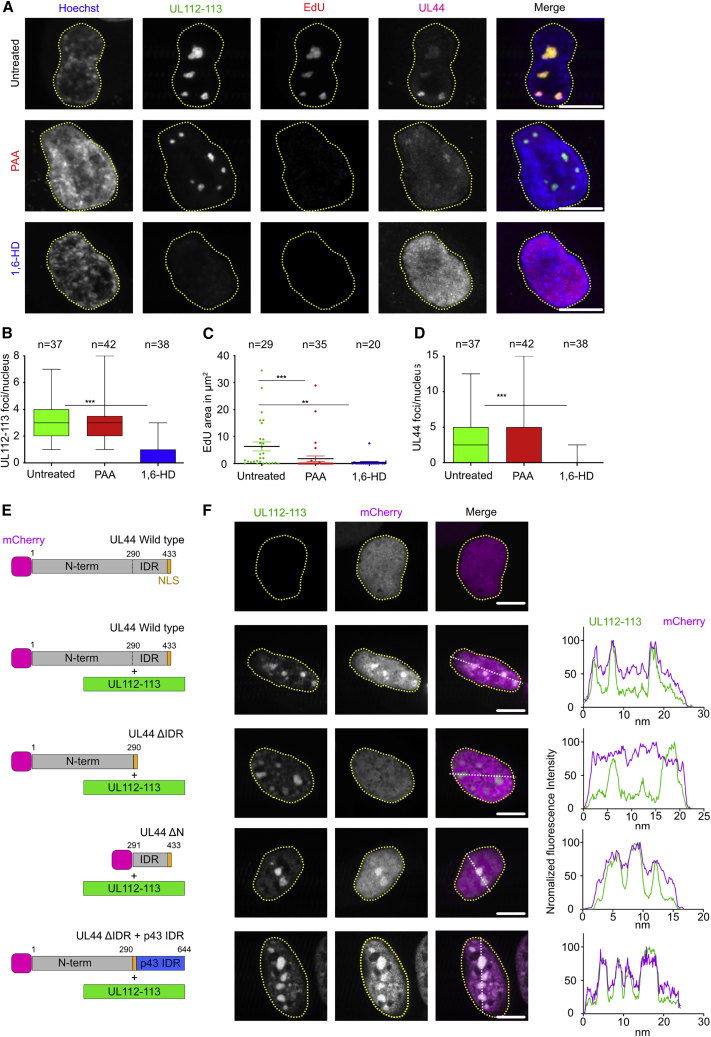
Figure 6.
Phase separation of UL112-113 facilitates recruitment of the viral DNA polymerase accessory factor UL44 to genomes
(A) Influence of inhibition of UL112-113 LLPS on genome replication. MRC-5 cells were infected with HCMV-mNeonGreen-UL112 (MOI = 1). At 24 hpi, cells were pulsed for 2 h with 10 μM EdU to label newly synthesized DNA. At the same time, cells were treated with PAA (250 μg/μL), 2% 1,6-HD, or left untreated. Cells were Click-labeled with AF555-Picolyl-azide (EdU), counterstained with Hoechst 33342, and UL44 was detected by indirect immunofluorescence.
(B–D) Influence of 1,6-HD on UL112-113 foci and genome replication. Number of UL44 positive foci, area of genome labeling, and number of UL44 positive foci under conditions as in (A). Only cells expressing UL112-113 and UL44 were analyzed. n = number of foci per nucleus analyzed.
(E) Schematic of mCherry-tagged UL44 constructs. UL44 consists of an ordered N-terminal region (aa 1–290), a disordered C-terminal IDR (aa 291–424), and an NLS (aa 425–433).
(F) HEK293A cells were co-transfected with UL44 and UL112-113 plasmids as indicated in (E) and imaged 48 h post-transfection. Intensity plots illustrate signal profiles along the white dotted lines. (A–D and F) The data shown are representative of three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01.